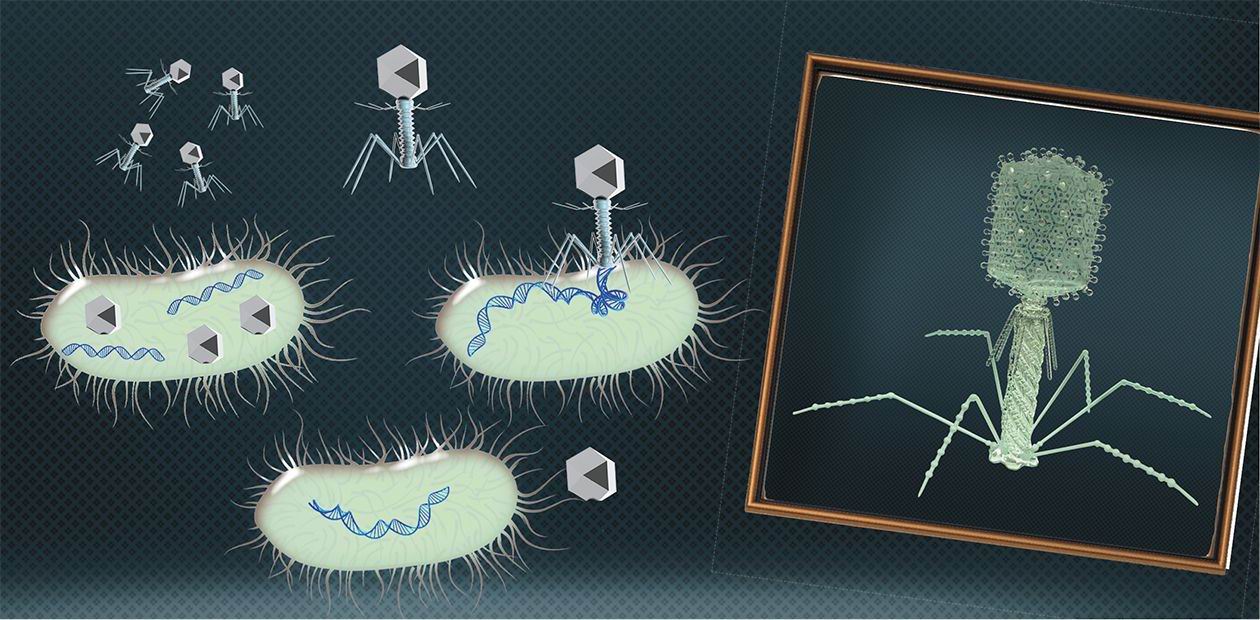
Bacteriophages: 100 years in the service of mankind
In the mid-last century, the science of biology made a leap forward when it established the molecular basis of living systems functioning. A crucial role in the successful research that resulted in determining the chemical nature of genetic molecules, breaking the genetic code and creating the gene manipulation technologies was played by the bacteriophages discovered at the beginning of the last century. Today, these bacterial viruses have learnt many jobs useful for people: they can be used not only as safe anti-infective drugs but also as disinfectants and even as basic elements of electronic nanodevices
When in the 1930s a group of scientists took up the problems of living systems functioning, their search for the simplest models lead them to bacteriophages – bacterial viruses extensively studied and applied in clinical practices. Indeed, there are no biological objects simpler than bacteriophages; in addition, they are easy to grow and analyze and have small genetic programs.
A phage is a minimal-sized natural structure that contains a densely packaged DNA or RNA genome and has nothing extra. It is encapsulated in a protein shell with a minimal set of devices designed to plant it into a bacterial cell. Bacteriophages cannot replicate on their own, and in this respect cannot be considered proper living objects. Their genes can only start working inside a bacterium with the help of its biosynthetic systems and stock of molecules necessary for synthesis. However, the genomes of these viruses are not fundamentally different from the genomes of more complicated organisms, therefore experiments on bacteriophages have allowed scientists to establish the guiding principles of the genome structure and work.
Subsequently, this knowledge and techniques developed as part of the study provided the basis for the development of biological and medical sciences as well as a wide range of biotechnological applications.
Fighting pathogens
The first attempts to use bacteriophages for treating bacterial infections were made almost directly after they were discovered, though the lack of knowledge and imperfect biotechnologies of the time prevented unconditional success. Nevertheless, further clinical practice showed that in principle bacteriophages could be successfully applied to treat gastrointestinal, urogenital and surgical infections as well as acute purulent-septic conditions.
Bacteriophages are our friends when we deal with bacteria pathogenic for humans. However, there are other, human-friendly bacteria employed by modern biotechnological productions as well as by conventional food processing, such as cheese making. Here, the phages can do a lot of harm since intensively growing large populations of microorganisms create favorable conditions for phage reproduction, which results in the lysis of industrial bacterial cultures. For cheese makers this is not a major concern because, as a rule, they use ferments consisting of many cultures, some of which can withstand the phage attack and maintain lactic fermentation. Serious difficulties emerge when just one specific bacterial strain is employed, for instance, in the production of antibiotics or therapeutic proteinsIn contrast to antibiotics, bacteriophages have a number of advantages: they have no side effects and are strictly bacterium-specific so do not disrupt the normal human microbiome. On the other hand, this high selectivity creates problems: a successful treatment requires an exact identification of the infectious agent and an individual selection of a bacteriophage.
Phages can be used for preventive purposes. For instance, the G. N. Gabrichevsky Moscow Scientific Research Institute for Epidemiology and Microbiology has developed FOODPHAGE, a prophylactic product made of a cocktail of bacteriophages to reduce the risk of acute enteric infections. Clinical research has shown that the administration of FOODPHAGE during a week allows the patients to get rid of the hemolyzing bacterium and other pathogenic and potentially pathogenic bacteria causing intestine dysbacteriosis.
Bacteriophages are suited for treating not only human infectious diseases but also these of domestic and livestock animals: mastitis in cows, colibacteriosis and colibacillosis in calves and pigs, salmonellosis in chicken… Phage drugs are especially easy to use for aquaculture – to treat farmed fish and shrimps – as they keep for a long time in the water. Bacteriophages can help to protect plants, too, though in this case the application of phage technologies is handicapped by exposure to sunlight and rain, which are destructive to the viruses.
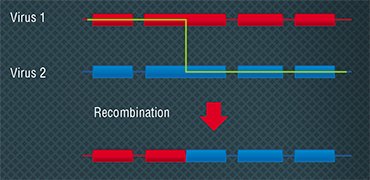
BACTERIOPHAGES AS FRUIT FLIES OF MOLECULAR BIOLOGY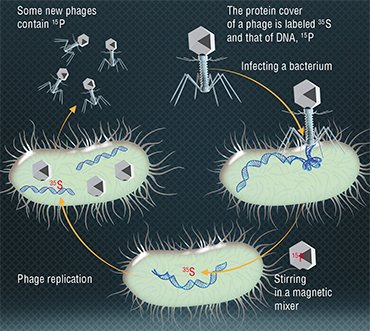 In 1946, at the 11th Symposium held in the famous Cold Spring Harbor Laboratory, the one gene- one enzyme hypothesis was enunciated. The bacteriologist A. Hershey and a “former” physicist, molecular biologist M. Delbrück reported an exchange of genetic traits between different phages when E.coli cells were simultaneously infected by them. This discovery made at the time when the physical carrier of a gene was not known suggested that recombination, a shuffling of genetic traits, is inherent not only in higher organisms but in viruses as well. This discovery made possible the later detailed study of molecular replication mechanisms. In the years that followed, experiments with bacteriophages allowed establishing the principles of generic programs’ structure and functioning.
In 1946, at the 11th Symposium held in the famous Cold Spring Harbor Laboratory, the one gene- one enzyme hypothesis was enunciated. The bacteriologist A. Hershey and a “former” physicist, molecular biologist M. Delbrück reported an exchange of genetic traits between different phages when E.coli cells were simultaneously infected by them. This discovery made at the time when the physical carrier of a gene was not known suggested that recombination, a shuffling of genetic traits, is inherent not only in higher organisms but in viruses as well. This discovery made possible the later detailed study of molecular replication mechanisms. In the years that followed, experiments with bacteriophages allowed establishing the principles of generic programs’ structure and functioning.
In 1952, A. Hershey and M. Chase showed by experiments that the genetic information of bacteriophage T2 is encoded, contrary to a popular belief, not in proteins but in DNA molecules (Hershey & Chase, 1952). The researchers observed reproduction in two groups of bacteriophages, one with radioactively labelled proteins and the other with DNA molecules. Bacteria were infected with these phages, and it turned out that only the viral DNA is transferred to infected cells, which proved the role of DNA in storing and transferring genetic information.
 In the same year, the American geneticists J. Lederberg and N. Zindler carried out experiments using two strains of Salmonella and bacteriophage P22. They showed that in the process of reproduction a bacteriophage could incorporate the DNA fragments of the host bacterium and transfer them to other bacteria infected by it (Zinder & Lederberg, 1952). This gene transfer from a host bacterium to a recipient was called transduction. The experimental results confirmed again the role of DNA in transferring information for inheritance.
In the same year, the American geneticists J. Lederberg and N. Zindler carried out experiments using two strains of Salmonella and bacteriophage P22. They showed that in the process of reproduction a bacteriophage could incorporate the DNA fragments of the host bacterium and transfer them to other bacteria infected by it (Zinder & Lederberg, 1952). This gene transfer from a host bacterium to a recipient was called transduction. The experimental results confirmed again the role of DNA in transferring information for inheritance.
In 1972, R. Beard and his colleagues, when investigating the replication (making a copy of cell information) of E.coli DNA, used bacteriophages as probes that can build into a bacterial cell genome and revealed that the process of replication goes in two directions along the chromosome (Stent, 1974)
Phages can be valuable for maintaining the microbiological safety of food products as antibiotics and chemical agents applied in food industry do not solve this problem. Moreover, they are not environmentally friendly. The scale of the issue can be demonstrated statistically: every year up to 40,000 people infected by salmonellosis are recorded in the USA and in Russia, and 1 % out of them die. The spread of this disease is largely connected with the farming, processing and consuming different kinds of poultry, and attempts to apply bacteriophages have produced promising results.
In 1969, A. Hershey, M. Delbrück and their colleague S. Luria were awarded the Novel Prize “for their discoveries concerning the replication mechanism and the genetic structure of virusesThus, the American company Intralytix makes phage products targeting Listeria, Salmonella and bacterial contamination. The have been licensed as additives preventing bacterial reproduction in foods; phages are sprayed over meat and poultry products, as well as over fruit and vegetables. Experiments have shown that a phage cocktail can be successfully applied in transporting and selling live pond fish to decrease the bacterial contamination of the fish and of the water.
An evident application of bacteriophages is disinfection, i. e. destroying bacteria in the places that should be clean of them, like hospitals and food processing facilities. For this purpose, the British company Fixed-Phage has developed a technology for fixing phage products on diverse surfaces, which keeps the phage biologically active for up to three years.
The seven days of Creation
Modern methods of synthetic biology allow not only modifying the phage genomes but also creating completely artificial active phages. This is not a big technological challenge: you only need to synthesize a phage genome and introduce it into a bacterial cell, and the genome on its own will start all the processes necessary to synthesize proteins and assemble new phage particles. In modern laboratories, this will take just a few days.
The first fully sequenced DNA-genome was the genome of the phage φ174, over 5,000 nucleotides long (Sanger et al., 1977). The sequencing was performed by the group of the English biochemist F. Sanger, the author of the DNA sequencing method of the same nameGenetic modifications serve to change the phage specificity and increase the efficacy of phage therapy. To achieve this, the most aggressive phages are fitted with recognition structures binding them with the target bacteria. In addition, virus genomes are imbedded with genes coding the proteins toxic for bacteria, which disrupt metabolism – such phages are more deadly for bacteria.
Bacteria have several mechanisms protecting them from antibiotics and bacteriophages; one such mechanism is the destruction of virus genomes by restriction enzymes targeting specific nucleotide sequences. The therapeutic potency of phages can be improved through “reformatting” their gene sequence at the expense of gene code degeneration in such a way as to minimize the number of nucleotide sequences “sensitive” to the enzymes, at the same time preserving their coding properties.
A universal method for protecting bacteria from any environment are the so-called biofilms, films containing DNA, polysaccharides and proteins and made jointly by some bacteria, which are proof against both antibiotics and therapeutic proteins. These films are pain in the neck for doctors as they facilitate the destruction of enamel, form on the surface of implants, catheters, and artificial joints as well as in airways, on the skin surface and so on. To deal with biofilms, special bacteriophages were designed containing a gene coding the specific lytic enzyme that destroys bacterial polymers.
ENZYMES “FROM BACTERIOPHAGE”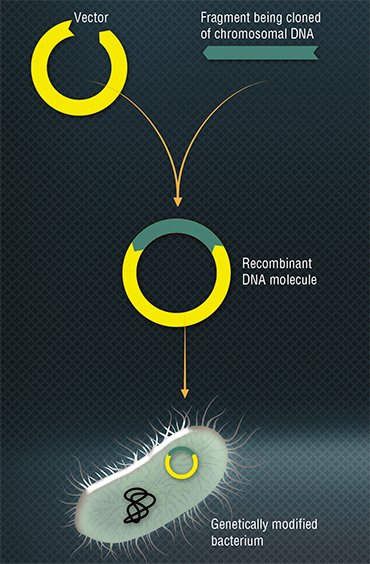 Research into bacteriophages resulted in the discovery of a great number of enzymes widely used today in molecular biology and genetic engineering. An example is restriction enzymes – a group of bacterial nucleases breaking down DNA. Back in the early 1950s, it was discovered that phages isolated from the cells of one bacterial strain tend to reproduce poorly in closely related strains. This meant that bacteria have a system for suppressing viral reproduction (Luria & Human, 1952). As a result, a restriction-modification system was found, a defense mechanism protecting bacteria against the invasion of alien DNA. The discovery of restriction enzymes (restriction endonucleases) supplied microbiologists with a priceless tool allowing manipulations with DNA: they learned to build some sequences into others and to cut the fragments needed, which ultimately led to the creation of recombinant DNA.
Research into bacteriophages resulted in the discovery of a great number of enzymes widely used today in molecular biology and genetic engineering. An example is restriction enzymes – a group of bacterial nucleases breaking down DNA. Back in the early 1950s, it was discovered that phages isolated from the cells of one bacterial strain tend to reproduce poorly in closely related strains. This meant that bacteria have a system for suppressing viral reproduction (Luria & Human, 1952). As a result, a restriction-modification system was found, a defense mechanism protecting bacteria against the invasion of alien DNA. The discovery of restriction enzymes (restriction endonucleases) supplied microbiologists with a priceless tool allowing manipulations with DNA: they learned to build some sequences into others and to cut the fragments needed, which ultimately led to the creation of recombinant DNA.
Another enzyme widely used in molecular biology is bacteriophage T4 DNA ligase, which joins the sticky and blunt ends of DNA and RNA double-stranded molecules. Recently, genetically modified variants of this enzyme have been developed that feature a higher activity.
Bacteriophages are the progenitors of most RNA ligases used in laboratory practices, which join single-stranded RNA and DNA molecules. In vivo, their main function is to repair broken RNA molecules. The most popular with researchers is bacteriophage T4 RNA ligase, which helps to attach single-stranded polynucleotides to RNA molecules in order to label them. The same technique applies to exploring the RNA structure, the binding sites of RNA and proteins, oligonucleotide synthesis, and so on. Newcomers to the class of routinely used enzymes are thermally stable RNA ligases extracted from bacteriophages rm378 and TS 2126 (Nordberg Karlsson, et al., 2010; Hjorleifsdottir, 2014).
Some polymerases, vitally important enzymes, have also been derived from bacteriophages. An example is the highly accurate phage T7 DNA polymerase, which has found application in various areas of molecular biology such as site-directed mutagenesis though it is mostly used to determine the primary structure of DNA.
The chemically modified phage T7 DNA polymerase was suggested as an ideal tool for sequencing DNA back in 1987 (Tabor & Richardson, 1987). A modification of this polymerase increased its efficiency by several times: DNA polymerization rate is higher than 300 nucleotides per second, so it can be used to amplify large DNA fragments. This enzyme became the predecessor of sequenase – a genetically modified enzyme used for Sanger sequencing. Sequenase is highly efficient and is able to include into a DNA sequence the nucleotide analogs used to improve the sequencing results.
Originating from bacteriophages are basic RNA polymerases (DNA-dependent RNA polymerases), enzymes catalyzing the process of transcription (reading RNA copies from a DNA matrix). Here belong SP6-, T7- and Т3-RNA polymerases called after the respective bacteriophages SP6, Т7 and Т3. These enzymes are used to synthesize in vitro antisense RNA transcripts, labeled RNA probes, etc.
Polynucleotide kinases catalyze the transfer of the phosphate group from the molecule ATP to 5’ end molecule of nucleic acid, exchange of 5’ phosphate groups or phosphorylation of 3’ ends of mononucleotides. The most commonly used in laboratory practices is the polynucleotide kinase of bacteriophage Т4. Its applications include experimenting with DNA labeling using phosphorus radioactive isotope, seeking restriction sites, DNA and RNA dactylography, and substrate synthesis for DNA or RNA ligases.
Molecular-biological experiments actively engage such bacteriophage enzymes as phage T4 polynucleotide kinase, commonly used for DNA labeling with radioactive phosphorous isotope, DNA and RNA dactylography, etc., as well as the DNA cleaving enzymes used to obtain single-stranded DNA matrices for sequencing and analyzing nucleotide polymorphism
The synthetic biology methods have helped to develop the bacteriophages equipped with most sophisticated weapons, which are used by bacteria against the phages themselves. These are CRISPR-Cas bacterial systems, which combine a DNA-cleaving nuclease and an RNA-sequence directing this enzyme towards a specific fragment of the virus genome. The “pointer” is a bit of the phage DNA that the bacterium keeps “for memory” in a special gene. When this protein-nucleotide complex discovers an analogous fragment inside a bacterium, it destroys it.
Having handled the principle of CRISPR-Cas operation, researchers tried to equip phages with a similar “weapon”: for this purpose, a complex of genes was introduced into the phage genome that codes the nuclease and RNA addressing sequences complementary to the specific sites of the bacterial genome. “Targets” can be the genes responsible for multiple drug resistance. The experiments were a complete success: the phages efficiently defeated the bacteria to which they had been “tuned.”
Phage antibiotics
Therapy does not necessarily require phages as such. Over millions of years of their evolution, bacteriophages have developed an arsenal of specific proteins – tools for recognizing the target microorganisms and manipulations with the victim’s biopolymers, based on which antibacterial drugs can be designed. The most promising proteins of this kind are the endolysin enzymes used by phages to destroy the cell wall when leaving a bacterium. These enzymes as such are powerful antibacterial substances not toxic for humans. Their efficacy and ability to hit the target can be improved by altering their addressing structures – proteins specifically binding with certain bacteria.
With respect to the cell wall structure, most bacteria fall into gram-positive (their membrane is covered with a very thick layer of peptidoglycans) and gram-negative (the peptidoglycan layer is between the two membranes). The application of natural endolysins is especially efficacious in gram-positive bacteria (staphylococci, streptococci, etc.) as their peptide-glycan layer is external. The gram-negative bacteria (Pseudomonas aeruginosa, salmonellas, colibacillus, etc.) are a target more difficult to hit since the enzyme has to penetrate the outer bacterial membrane in order to approach the internal peptide-glycan layer.
To deal with this problem, the so-called artilysins were created – modified variants of natural endolysins containing polycationic or amphipathic peptides, which destabilize the outer membrane and deliver endolysin directly to the peptide-glycan layer. Artilysins feature a high germicidal activity and have proved efficacious in treating otitis in dogs (Briers et al., 2014).
An example of modified endolysin selectively targeting certain bacteria is the drug P128 developed by the Canadian company GangaGen, Inc. It is a biologically active fragment of endolysin combined with lysostaphin – an addressing protein molecule binding with the surface of staphylococci cells. The fusion protein obtained is highly active against a variety of staphylococcus strains including these with multiple drug resistance.
Bacteria “counters”
In addition to being diverse therapeutic drugs and “disinfectants,” bacteriophages are valued by microbiologists as convenient and precise analytical tools. For instance, owing to their high specificity they are natural analytical reagents capable of detecting the bacteria of a certain species and strain.
In the easiest version of this investigation, diverse diagnostic bacteriophages are added, drop by drop, in the Petri dish with a nutritional medium seeded with a bacterial culture. If a bacterium turns sensitive to a phage, the bacterial “lawn” develops a “patch” – a transparent area with killed and lysed bacterial cells.
Analyzing phage replication in the presence of target bacteria, we can evaluate the number of the latter. Since the number of phage particles in the solution increases in proportion to the number of the bacterial cells it contains, to evaluate the number of the bacteria we need only to determine the titer of the bacteriophage.
The specificity and sensitivity of this analytical reaction is quite high; the procedures are easy to carry out and do not require any sophisticated equipment. Importantly, the bacteriophage-based diagnostic systems signal the presence of a living pathogen itself whereas other methods, such as PCR and immuno-analytical, show only the presence of the biopolymers belonging to this bacterium. The diagnostic methods of this kind are especially convenient to use in ecological research as well as in food industry and agriculture.
Today, to detect and estimate the number of the various strains of microorganisms, special reference species of phages are used. Very fast, functioning virtually in the real-time mode, analytical systems can be created on the basis of genetically modified bacteriophages, which, on entering a bacterial cell, launch the synthesis of fluorescent (or capable of luminescence) reporter proteins, such as luciferase. When the necessary substrates are added to such a medium, it will generate a luminescent signal with the intensity proportional to the content of bacteria in the sample. These “light-labeled” phages have been developed to detect dangerous pathogens – agents of plague, tuberculosis, as well as of plant infections.
Probably, modified phages will help to deal with an old problem of global importance: the development of cheap and rapid techniques for detecting tuberculosis agents at an early stage of the disease. This is a big challenge as the mycobacteria causing tuberculosis grow very slowly when cultivated in laboratory conditions. Therefore, conventional diagnostics may take a few weeks.
Phage technology speeds up this task. The point of the technology is adding the bacteriophage D 29 targeting a wide range of mycobacteria to the blood samples being analyzed. After that, the bacteriophages are separated, and the sample is mixed with a fast-growing non-pathogenic mycobacterial culture, which is also sensitive to this bacteriophage. If the blood originally had mycobacteria infected with phages, the new culture will also produce the bacteriophage. In this manner, single mycobacterial cells can be detected, and the diagnostics takes just 2—5 days instead of 2—3 weeks (Swift & Rees, 2016).
Phage display
Today, bacteriophages are widely applied as simple systems to produce tailor-made proteins. We are talking about phage display – a most efficient molecular selection technique developed in the 1980s. The term was coined by an American, George Smith, who proved that Escherichia coli phages could be used to produce a viable modified virus with a foreign protein on its surface. To accomplish this, an appropriate gene is inserted into the phage genome, which merges with the gene coding a surface virus protein. These modified bacteriophages can be isolated from a mixture of wild-type phages owing to the “foreign” protein’s ability to bind with specific antibodies (Smith, 1985).
From Smith’s experiments two important conclusions followed: first, using the recombinant DNA technology, you can generate a vast variety of populations numbering 106—1014 phage particles, each of them carrying on its surface different variants of proteins. These populations were called combinatorial phage libraries. Secondly, a specific phage (for example, a phage able to bind with a certain protein or an organic molecule) isolated from a population can be replicated in bacterial cells to produce an infinite number of descendants with given properties.
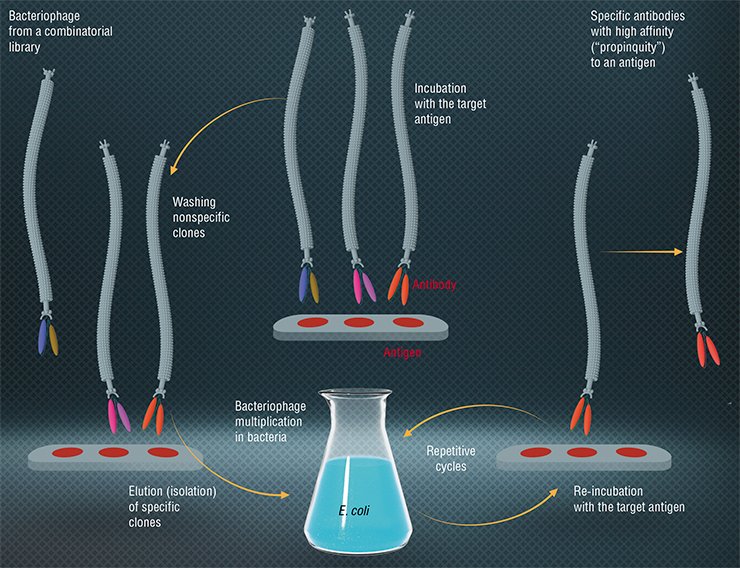
Today, phage display is used to make proteins that can bind selectively with therapeutic targets; for instance, the proteins exposed on the surface of the phage M13 are able to recognize and interact with tumor cells. The function of these proteins in a phage particle is to “pack” nucleic acids; therefore, they are well suited to produce gene therapy drugs, only in this case they form a particle with a therapeutic nucleic acid.
The two main current application areas of a phage display are the peptide-based technology and protein-based and domain-based technology. The former is used to study receptors and to map the antibody-binding sites, to create immunogens and nanovaccines as well as to map the substrate-binding sites in proteins-enzymes. The protein-based and protein domain-based technology helps to select antibodies with given properties, to study protein-ligand interactions, and to screen the expressed fragments of a complementary DNA and directed protein modifications.
Phage display can be used to introduce recognition groups into all species of surface virus proteins as well as into the main protein forming the bacteriophage body. The introduction of peptides with given properties into surface proteins gives you a whole range of valuable biotechnological products. For example, if this peptide imitates the protein of a dangerous virus or bacteria recognized by the immune system, the modified bacteriophage is a vaccine that can be produced easily, quickly and safely.
If you “address” the end surface protein of a bacteriophage to cancer cells, and add reporter groups (e. g., fluorescent or magnetic) to another surface protein, you will obtain a tumor detector. And if you add a cytotoxic drug to the particle (which can be done easily with the help of modern bioorganic chemistry), you will get an anti-tumor drug.
An important application of the protein phage display technique is the creation of the phage libraries of recombinant antibodies, where the antigen-binding fragments of immunoglobulins are placed on the surface of phage particles fd or M13. Of special interest are the libraries of human antibodies because these antibodies can be used in therapy without limitations. In the recent years, the pharmaceutical market of the USA alone has offered more than a dozen of therapeutic antibodies developed using this method.
“Industrial” phages
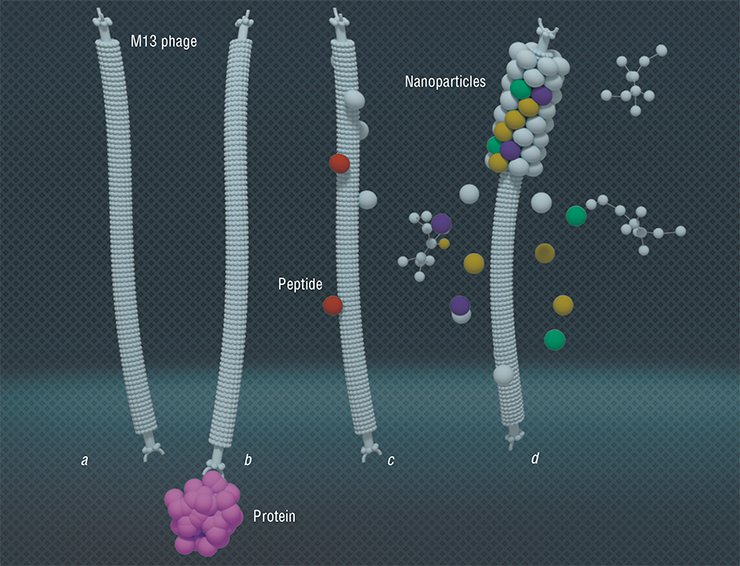
Along with the uses described above, the phage display technique has found a completely unexpected application. Bacteriophages are primarily nano-sized particles of a certain structure with proteins located on their surface. With the help of phage display, the phages can acquire the property of binding specifically with the necessary molecules. These particles offer a widest range of opportunities to create materials with a given architecture and smart molecular nano-devices using eco-friendly production technologies.
Technologies allowing arranging filamentous bacteriophages one by one (tail-to-tail) have been developed. The resulting multiphage structures are ordered nano-matrices that can be applied to make transistor and diode devices
Since the virus is a rigid structure governed by a certain dimensional equation, it can be used to make porous nano-structures with a given surface area and the necessary distribution of pores within its structure. As is known, the surface area of a catalyzer is a critical parameter defining its efficiency. Current technologies for forming a thinnest layer of metals and their oxides on the bacteriophage surface make possible catalyzers with an extraordinarily developed regular surface with the dimensions prescribed. (Lee et al., 2012).
The researcher from the Massachusetts Institute of Technology A. Belcher used bacteriophage M13 as a template for growing rhodium and nickel nanoparticles and nanowires on the surface of ceric oxide. The catalyst nanoparticles obtained encourage the conversion of ethanol to hydrogen – in this manner, this catalyst may prove valuable for modernizing the existing hydrogen fuel cells and generating new ones. As compared with an analogous “conventional” catalyst, the catalyst grown from a virus template has a higher stability and is less prone to ageing and surface deterioration (Nam et al., 2012).
The coating of filamentous phages with gold and indium dioxide has produced electrochromic materials – porous nanofilms changing color with a change of the electric field 1.5 times quicker than the analogues known. Such materials are promising for making energy-conserving ultrathin displays (Nam et al., 2012).
In the Massachusetts Institute of Technology, bacteriophages have become the basis for the production of powerful and exceedingly compact batteries. For this purpose, living and genetically modified phages M12 were used, which are benign for people and are able to attach to their surface the ions of various metals. The self-assembly of these viruses resulted in the structures of a preset configuration, which, when coated with metal, formed sufficiently long nanowires that became the basis for the anode and cathode. The self-formation of the anode material employed the virus capable of attaching gold and cobalt oxide, and in the case of the cathode material, it was a virus capable of attaching iron phosphate and silver. The final phage also had the ability to “connect” the ends of the carbon nanotube, which is necessary for an efficient transfer of electrons.
In addition, bacteriophage M13, titan dioxide and single-wall carbon nanotubes have served to create materials for solar batteries (Dang et al., 2011).
The recent years were marked with extensive studies of bacteriophages, which find themselves new applications not only in therapy but also in bio- and nanotechnologies. An evident practical result should be the emergence of a new large area of personalized medicine as well as the development of a wide range of technologies in food industry, veterinary medicine, agriculture and production of modern materials. We expect that another hundred years of bacteriophage research has at least as many discoveries in store as the first one.
References
Bacteriophages: biology and application/Ed.: E. Cutter, A. Sulakvelidze. Moscow: Nauchnyi mir. 2012.
Mc Grath S., van Sinderen D. Bacteriophage: Genetics and Molecular Biology. Horizon Scientific Press, 2007.
Stent G., Calendar R. Molecular Genetics. Moscow: Mir. 1974. 614 p.
Tikunova N. V., Morozova V. V. Phage display based on filamentous bacteriophages: application for the selection of recombinant antibodies // Acta Naturae. 2009. N. 3. P. 6–15.