Catalytic RNA:
Drug from the Prebiotic World
Once considered marginal, the concept of the RNA world is now firmly established in the thinking of biologists. Born in the 1960s and then founded in an extensive body of work on the catalytic and information storage properties of RNA in the 1980s, the hypothesis states that at some stages of prebiotic evolutions the early replicators, or entities that multiply and compete for limited resources, were made exclusively of RNA. Proteins and DNA are merely later important additions to the scheme of information transfer that allowed it to run more smoothly. The SB RAS Institute of Chemical Biology and Fundamental Medicine (Novosibirsk) has long been involved in basic and applied research on various RNA species. In 2013, the institute received a mega-grant from the Russian government for further development of this research area under the leadership of Prof. Sidney Altman (Yale University, United States), a Nobel Prize winner for the discovery of the catalytic properties of RNA
We can observe supposedly many relicts from those enigmatic times – RNA molecules that do something more in the cell than conveying the genetic information from DNA to protein. Many regulatory, catalytic, adaptor and scaffold RNA molecules show us how pre-life could once be organized.
One of such remnants for the past forty years went from a biochemical curiosity to—possibly—something as close to a panacea remedy as it could be. Its story is an excellent illustration of how purely blue-sky research, through detailed understanding of the working mechanism of something, can bring forward a new creative idea that could possibly transform a whole field of human enterprise (medicine in this case). Our “something”, RNase P, is intimately involved as an accessory of post-transcriptional mechanisms.
RNA for dummies
According to the central dogma of molecular biology, translation is a process that transfers the text written in RNA letters (e.g., …AAAUUUCGAUC…) into another text written by amino acid letters (e.g., …YALE… where Y is the amino acid tyrosine, A is alanine, L is leucine, and E is glutamic acid). Translation occurs in protein-making factories called ribosomes. Not only do they make protein from coding RNA, but also their function is ultimately based on two classes of non-coding ones, ribosomal RNA (rRNA) and transfer RNA (tRNA). To be exact, the ribosomes of Escherichia coli, this workhorse of molecular biology, contain three molecules of rRNA, known as 16 S, 23 S and 5 S rRNA, while we humans have four in our ribosomes: 18 S, 28 S, 5.8 S and 5 S. To make a biologist’s life somewhat more complicated, in E. coli there are seven genes for any of the rRNAs, which code for products very similar but nevertheless diverging at a few places. The situation in humans is, speaking broadly, similar.
When we move to tRNA, things become even more complicated, since E. coli has 86 genes coding for the set of 20 standard tRNA, and of course humans are more complex with 497 nuclear and 22 mitochondrial tRNAs. Obviously, this is far more tRNAs than suffice to code for 20 amino acids, or even 61 coding triplets. Some of this redundancy is functional, allowing the cell to use certain tRNAs under defined circumstances.
tRNA structure is often drawn as a cloverleaf. A remarkable structure, considering the small size of the molecule: most tRNAs fall between 75 and 95 nucleotides long. The cloverleaf consists of a “stem” and three loops. One is the anticodon loop, thus called because it contains the antocodon, the codon-recognizing stretch of three nucleotides. Two other loops are named after modified bases they carry: D-loop contains a dihydrouracil residue, and TC-loop, a pseudouridine residue, neither of which is normally found in coding RNAs. They do not exhaust the list of deviant nucleobases found in tRNA: inosine, ribothymidine, 4-thiouracil and a panoply of others all make their way into mature tRNA.
In addition to the main loops, a “variable loop” is present; its length is one of the diagnostic features for tRNA classification: Class I molecules have short loops (3-5 nucleotides) while in Class II molecules, the variable loop may exceed all main loops. The acceptor stem contains a short 3’-terminal flap, usually 4 nucleotides long, ending in …CCA, which will then covalently bind the amino acid to form aminoacyl-tRNA, the main construction block for protein synthesis.
With the seemingly iconic status of the cloverleaf, the actual 3D structure of tRNA does not resemble it at all, being folded into an L-like shape approximately ~60×60 Å, with the anticodon and the 3’-end of the acceptor loop are separated in space as far away from each other as possible.
Indiscriminate ribozyme
tRNA is usually made as a long precursor molecule. Besides modification of the bases to create dihydrouracil, pseudouridine, etc. a tRNA can be split off a longer precursor transcript containing several other genes (in bacteria, several adjacent genes are usually transcribed as a single unit). An enzyme, RNase E, releases the tRNA precursor from many such transcripts, yet the tRNA has extra nucleotides both at the 5’ and in the 3’ side. Several specific nucleases then trim the pre-tRNA to its final form.
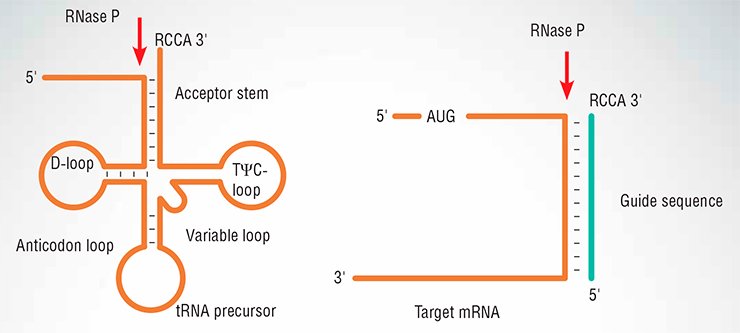
One of these nucleases, RNase P, has a job of cleaving pre-tRNA to expose the 5’-end of the mature molecule, just next to the …CCA 3’-tail in the cloverleaf. RNase P has been at the center of my lab’s work since the early 1970s, when I discovered that tRNA is made from a precursor. Of course, the most startling and unexpected finding about RNase P was reported in our 1983 Cell paper – the enzyme turned out to be a catalyst based on RNA.
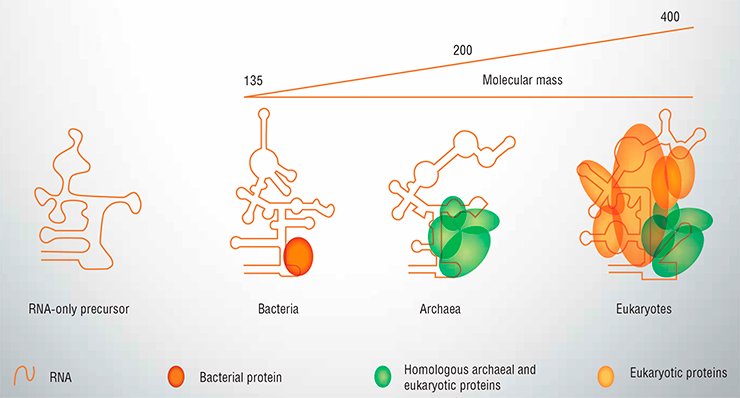
Actually, the catalytic RNA in RNase P never works alone. It can, in a test tube, but in all living organisms it is found in a complex with at least one protein. In bacteria, RNase P contains only one protein, RnpA. In Archaea, unicellular prokaryotes that are closer to eukaryotes than to bacteria, RNA associates with 4-5 proteins to form RNase P, none of which is related to RnpA. In eukaryotes, there are up to ten proteins bound to the catalytic RNA, some of them are homologous to the archeal ones while others are unique for eukaryotes (Fig. 1). Although the RNA part retains some catalytic activity even when purified, it cannot work in the cell lacking the protein component of RNase P. Yet it is quite reasonable to believe that all today’s RNases P ultimately evolved from an RNA-only precursor, whenever it existed – in the days of the RNA world or later when the machinery of protein biosynthesis started to assume its present form.
Through our studies of the mechanism of RNase P action we have defined the minimum requirements for its substrate. In other words, we have found what the RNA precursor must look like to be cut by RNase P.
Surprisingly, the cloverleaf structure is not needed at all. RNase P efficiently cleaves any RNA provided it resembles a tRNA stem – is partially double-stranded near its 3’-terminus, with the RCCA flap (R is either adenine or guanine) dangling free. RNase P would cleave such a substrate at its 5’-side at a junction with the flap (Fig. 2). Even if we have two RNAs, one of any sequence (target), another complementary to it plus the flap (external guide sequence), the target would be cut.
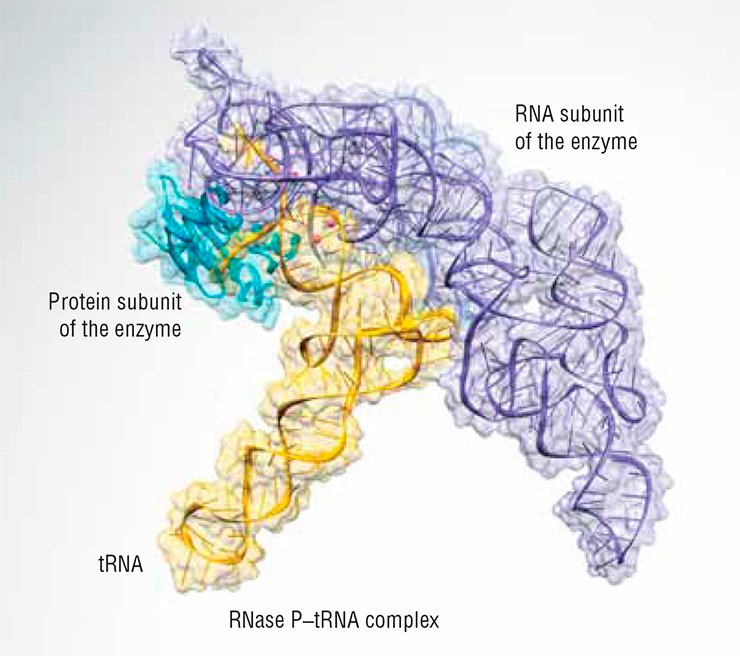
The X-ray structure of RNase P in a complex with tRNA clearly shows why this is so (Fig. 3). The ribozyme binds its substrate mostly through shape complementarity. Specific bonds of Watson–Crick type are only made near the 3’-terminus where the invariable flap is located. Moreover, even chemically altered RNA guide sequences can be used if they fulfill the same criteria of target recognition.
Combating drug resistance
The long-sought explanation of RNase P substrate specificity was at the same time the starting point of another quest. If, as it turns out, we can cleave any RNA with RNase P, why not try to turn it into a weapon to eliminate unwanted RNA? If, for example, we could introduce an external guide RNA into a bacterial cell, the bacteria’s own RNase P would destroy the target RNA recognized by the guide RNAs in the cell – preferably some important pathogenic RNA, the loss of which would be lethal.
 Why bother? The reason is clear. The widespread uncontrolled use of antibiotics caused a rise in the occurrence of drug-resistant microbes. Bacteria can easily exchange pieces of genetic material, called plasmids, and plasmids very often carry genes that quickly metabolize drugs, so if such a plasmid appears in a bacterial population, or even in a bacterial ecosystem containing different species, pretty soon all the cells will pick it up provided that there is an antibiotic around.
Why bother? The reason is clear. The widespread uncontrolled use of antibiotics caused a rise in the occurrence of drug-resistant microbes. Bacteria can easily exchange pieces of genetic material, called plasmids, and plasmids very often carry genes that quickly metabolize drugs, so if such a plasmid appears in a bacterial population, or even in a bacterial ecosystem containing different species, pretty soon all the cells will pick it up provided that there is an antibiotic around.
Another way to develop resistance is to mutate a protein target for the drug, or produce more of this protein, or activate genes responsible for pumping the drug out of the cell. The scale of the problem and the associated risks are tremendous. For example, multi-drug-resistant tuberculosis, insensitive to the first-line drugs isoniazid and rifampicin, now accounts for 4% of new TB cases and 20% of the recurrent cases worldwide, and has been reported in all countries. If the bacteria additionally develop resistance to quinolone antibiotics and second-line drugs such as kanamycin, they cause extremely drug-resistant tuberculosis, which numbers in tens of thousands cases per year. What’s more, India, Iran, and Italy have reported cases of totally drug-resistant tuberculosis, not responding to all first- and second-line antibiotics.
 To make matters worse, development of a new antibiotic costs millions of dollars, and companies are not exactly queuing in line to spend these dollars and to get rewarded by low-cost, low-need products. Patients would spend tens of thousands of dollars for a cancer chemotherapy course that prolongs life for a few months, but are reluctant to pay $100 to cure an infection.
To make matters worse, development of a new antibiotic costs millions of dollars, and companies are not exactly queuing in line to spend these dollars and to get rewarded by low-cost, low-need products. Patients would spend tens of thousands of dollars for a cancer chemotherapy course that prolongs life for a few months, but are reluctant to pay $100 to cure an infection.
Perfect hit
Realizing the potential of RNase P external guides as drugs, we first found a way to deliver them efficiently in the bacteria. We conjugated the guides with cell-penetrating peptides, short protein fragments that can either create a hole in a bacterial cell wall or be actively transported into a cell. The guide thus enters the cell as a passenger.
Also, we decided to use guide sequences with a non-natural chemistry: not RNA or DNA, but so-called morpholinos, in which a six-member morpholine heterocycle substitutes for the sugar ring of natural nucleic acids. Morpholinos are more stable in cells and produce tighter complexes with their target nucleic acids.
As a target, we chose mRNA made from gyrA, a gene coding for DNA gyrase, an enzyme critically important for bacterial replication. Gyrase is a target for many important antibiotic groups, such as fluoroquinolons (including the well-known agent ciprofloxacin) and aminocoumarins, and its elimination is lethal. Notably it is possible to find a guide sequence common for many pathogens in the sequence of gyrA. We have tested the bactericidal properties of our guide sequences in several bacterial species, either pathogenic themselves or models of serious pathogens. In most cases, the viability of the cells was much less than 1%, although the concentrations of the drug used in these experiments were quite high in comparison with antibiotics in use today.
Encouraged by this first success, we turned our attention to malaria. Plasmodium falciparum, one of the causative agents of malaria, is not a bacterium but a unicellular eukaryote. Yet the resistance to most anti-malarial drugs is also a problem in clinical isolates. When we designed a guide sequence to target cleavage of DNA gyrase mRNA, and introduced it into erythrocytes infected with P. falciparum, the parasite ceased to grow – but it thrived if the guide was to bacterial gyrA, showing that the targeted mRNA is destroyed very specifically.
An unfortunate fate of a great many new drugs ends when they fail to perform in vivo despite showing promising activity in preliminary tests. Thus, we tried our approach in a real-life situation: infected wounds. Of course, these were not human wounds; mice served as a model. Their backs were punctured with a special device to make a small wound, which were infected with Staphylococcus aureus, a usual bacterial cause of skin infection in humans. The followng day, we covered wounds with a gel that contained either the guide morpholino directed against S. aureus gyrA and confirmed to work in vitro on both wild-type and antibiotic-resistant strains, a non-specific morpholino, or none at all. In treated mice, the wound healed much more quickly than in the control groups. Under the microscope, we saw that the treatment caused better regeneration of epithelium and the mature collagen layer. What is more, the bacteria, which still lingered in the wounds, were much less abundant in treated lesions than in untreated ones. The strict test of efficiency in a mammalian non-human organism was passed at least in the backs of mice.
Very importantly, the RNase P-based strategy promises to overtake microbes in the race once believed lost by humanity, that of new drugs development vs drug resistance development. When a new drug costs about 100 million dollars and takes ten years from conception to market, and then resistance arises in a couple of years free of charge for the bacteria, the winner seems clear. Guide sequence-dependent RNA cleavage may turn the tables. Once the safety and efficiency for a general group of drugs, say morpholinos with a particular cell-penetrating peptide, is established, we may quite freely vary the sequence and target various essential genes of the infectious agent, or use combinations of guides to different targets, making resistance development close to impossible. And, extremely unusual for pharmacology, the same type of drug can act on bacteria, protists, fungi, perhaps even on viruses and particular cancer cells, if we can deliver the guide molecules to the appropriate targets.
So why do doctors still rely on ampicillin, chloroquine and the like to treat infections? The answer is mostly economic. Morpholino guides (or guides made on any other chemical basis) are indeed more expensive than mass-produced small-molecule antibiotics, and companies are simply afraid that patients, and more importantly, doctors, will prefer to buy old-generation drugs. However, they are not dramatically more expensive: a dose would cost anywhere from 2 to 3 dollars more than a regular antibiotic. Given high efficiency and less complications of guide-based therapy, it seems that time has come for scientists, clinicians, companies, and regulatory bodies to make an investment that can bring back a great reward.
Prof. D. Zharkov kindly produced this summary of the lecture given at Novosibirsk, May 2014. The funding of the joint laboratory in Novosibirsk was provided by the Russian Ministry of Education and Science.