Parasitosis from A to T
Perhaps, the large-scale project aimed at a complete genomicsand proteomics-based decoding of Opisthorchis felineus, launched at the Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, will not only implement its minimum program—to understand the nature of the parasitosis caused by this agent and reliably control it but also fill the almost complete vacuum in the genomic research in Russia
Historically, studies in the field of experimental biology have followed the same developmental logic. The first stage was accumulation of experimental data on a certain biological system, frequently mosaic and sometimes controversial, which was due to several factors—differences in the experimental approaches, data processing techniques, and other less evident yet not less important reasons. The accumulated body of data allowed the corresponding biological phenomenon to be pondered, the apparent controversy of the data from various sources to be eliminated, and a theoretical model to be constructed. Based on such a model, new research directions were planned to clarify the yet unclear sides of the phenomenon and determine the factors providing for regulation and control of the system. This was succeeded by the next stage of data accumulation, and the cognition cycle was reproduced at a new level.
The current state of Russian genomic research dooms Russian biology, medicine, and biotechnology to the position of world outsidersThe eve of this century opened a new era for the basic and applied biological research. The level of advances in biotechnologies and bioinformatics (mathematical and information support of research) made it possible to turn from a mosaic acquisition of information in odd experiments to a systemic approach, which was implemented in genome sequencing projects. Indeed, the overall information about a biological species is stored in the genomic DNA in a four-letter code, and decoding such a genetic text gives the most comprehensive, and what is most important, systemic information about a living organism and the mechanisms that determine its structure, development, and function.
A genomic outsider
The end of the last and the beginning of the current century gave birth to a constellation of genome sequencing projects aimed at decoding genomes of various complexities—from viral and bacterial to eukaryotic. These projects contribute to various spheres of human activity—medicine, agriculture, biotechnology, and basic science. It is assumed that a future decrease in the cost of genome sequencing will make it possible, for example, to sequence individual declining species with an aim to preserve biological diversity.
The range of scales of genome sequencing projects is amazing: from a bacterial genome containing a single chromosome to the global initiative of the US Department of Energy—the Microbial Genome Project, where 485 microbial genomes have been deciphered and the genomics of 30 microbiological communities have been studied. The organizers of this global project intended that acquisition of detailed genomic information about the microbial communities (as these particular organisms constitute the major part of the planetary biomass) form the background for remediation of water bodies and lands, control of pathogenic microorganisms, and other activities providing sustainable development of the planetary ecosystem.
Unfortunately, our country is far behind this world trend. In 2007, the Institute of Physicochemical Medicine, Russian Academy of Sciences, completed the sequencing of the first “Russian” bacterial genome, Acholeplasma laidlawii, its computer annotation, and proteomic profiling; and the Bioengineering Center, Russian Academy of Sciences, is soon to complete the sequencing of the genome of a psychrophilic (cold-loving) bacteria.
GENOMIC SEQUENCING PROJECTS are directed to eventually determine the complete genomic sequence of an organism, be it a virus, bacterium, fungus, plant, or animal. For this purpose, it is necessary to find out the sequence of the DNA constituting each chromosome of a particular species. Thus, we need “to read” only one chromosome for a bacterium versus 22 autosomes pairs and two sex chromosomes for humans. The most famous and largest project in this field was the Human Genome Project, commenced in 1990. Sequencing the smallest human chromosome, chromosome 22, was completed in 1999, and publication of the sequence of the longest human chromosome, chromosome 1, in 2006 completed the Project.Historically, the first genomic approach was partitioning the genome into overlapping segments (mapping), sequencing these segments, and “assembly” of the complete genome. Later Venter and Smith (Nobel Prize laureates for the discovery of restriction endonucleases) developed the shotgun sequencing technique, when genomic DNA is fragmented by ultrasound; the sequences of these fragments are determined; and the complete genome is reconstructed from these fragments with the help of specialized software.
Genome projects comprise the following stages:
(1) Preparation of genomic DNA, its fragmentation, and sequencing (with the help of automated sequencers, which determine the sequences of DNA regions with a length of 900 letters, nucleotides).
(2) “Assembly” of the genome, i.e., the restoration of DNA sequences in each chromosome of a species from the sequenced fragments. This is not a simple task, made even more intricate by the presence of numerous identical sequences, the so-called repeats, in the genome. Specialized software packages are developed for this purpose.
(3) Genome annotation is the process of attaching biological information to sequences. It consists of two main steps: identifying elements of the genome (gene finding) and attaching biological information to these elements (gene structure, regulatory and coding regions, biological and biochemical functions of the protein products of genes, regulation of gene expression, and interactions with other genes). Genome annotation is performed both with the help of specialized software and manually using the experience of the experts in genome deciphering.
Upon sequencing the main body of information contained in the genome, there are often some regions left difficult to sequence (usually associated with DNA repeats); at this stage, the sequencing results are called the working draft sequence. As the sequencing process is sometimes not free from mistakes, it can require re-sequencing of genome fragments.
Many genome projects are not confined to a mere “reading” of the entire chromosomal DNA and also include transcriptomics and proteomics components, i.e., study of the structure of RNA transcripts and corresponding protein products, which lifts the project to a new functional level.
Choosing a biological species for genome sequencing is determined by several factors: the sequencing of humans and popular genetic models (the fruit fly Drosophila melanogaster, nematode Caenorhabditis elegans, bread mold Neurospora crassa, bacterium Escherichia coli, and so on) were initiated first. A number of projects are aimed at sequencing the pathogens menacing human health: viruses, bacteria, and fungi. It is not surprising that the first completed genome projects were virus projects just due to their small size. Several virus genomes (in particular, equine encephalomyelitis, Ebola, Marburg, Crimean hemorrhagic fever, and other viruses) were sequenced for warfare purposes; recently, the genome of the virus causing atypical pneumonia (SARS) was sequenced in very short terms in response to the demand of medicine. Archaebacterial genomes are sequenced as these microorganisms are able to inhabit most extreme ecological niches—the ocean bed, methane fields, natural hot springs, etc. In these projects, the focus is on the genes determining the corresponding abilities.
Important agricultural plants—rice (Oryza sativa), wheat (Triticum aestivum), and corn (Zea mays)—are also covered by genome projects. An interesting project performed under the International Grape Genome Program is aimed at improving the wine quality and searching for the genetic determinants of wine taste. Another reason for sequencing genomes is that the acquired information can clarify key issues of biological evolution. For example, the Chimpanzee Genome Project shed some light on the human evolution, as several genes involved in the human speech development were discovered and their distinctions from the corresponding chimpanzee genes allowed the scientists to infer that these genes had evolved due to the selection connected with human speech behavior.
As for the the higher eukaryotes, the sequencing of their genomes in this country is in an embryonic state. Participation of some Russian institutions in the genomic research performed abroad does not solve the problem. If the situation is not resolved in the nearest future, the Russian biology, biomedicine, and biotechnology will be doomed to yield not only to the world leaders, but also to the known outsiders of the world biological science.
Perhaps, the large-scale project aimed at a complete genomics and proteomics decoding of Opisthorchis felineus, launched at the Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, will not only implement its minimum program—to understand the nature of the parasitosis caused by this agent and reliably control it—but also fill the almost complete vacuum in the genomic research in Russia.
Parasite in focus
Even the minimum program for this project covers a wide range of objectives and should give results with a high potential of practical application. Indeed, opisthorchiasis is a most dangerous disease, prevalent almost exclusively in Russia and the CIS countries. Moreover, West Siberia, where the Novosibirsk oblast is located, is intensive natural focus of this parasitic disease with a very high infestation rate of both the human population and many species of domestic and wild animals.
Decoding the genetic text of genomic DNA gives comprehensive and systemic information about a living organism and the mechanisms determining its structure and vital activitiesThe problem of sanitary and epidemiological monitoring of this focus as well as diagnosing this disease in patients is further complicated by the absence of simple and reliable diagnostic test kits. The presence of several liver fluke species in the focus (it has been proved that at least one of these species, Metorchis bilis, can parasitize humans) adds complexity to the epidemiological and clinical picture of this parasitic disease and requires elaboration of the test kits that would combine a high sensitivity with the ability to precisely diagnose the species of liver parasites.
The currently available drugs for treating opisthorchiasis are rather toxic, while the incompleteness of the biological data on the morphology of liver fluke, the structure and functions of its genes, and genetic control of its lifecycle hinders the development of new antihelminthic drugs and vaccines. The situation is well illustrated by the fact that before the pilot phase of this project, liver fluke morphology was described only at the level of light microscopy, i.e., at the level of methods developed before the mid-last century.
In addition to these practical problems, O. felineus is of exceptional interest for various fields of basic science. In particular, it is still vague for ecologists why a large freshwater body, the Malye Chany Lake in the Novosibirsk oblast, based in the largest opisthorchiasis focus remains uninfected. Geneticists are intrigued by the phenomenon that this parasite is able to reproduce not only in the adult but also in the larva stage! This is a very rare natural phenomenon (in addition to liver flukes, it has been described for some beetles, marine crustaceans, and several other species). Thus, one liver fluke larva at an initial stage of its lifecycle produces a hundred of genetically identical copies—a real large-scale cloning. However, the corresponding molecular genetic mechanisms are still a mystery.
The problem of opisthorchiasis
Thus, the focus of this project is the liver fluke (cat/Siberian liver fluke) O. felineus, which is a lower multicellular animal. As the agent of human opisthorchiasis, O. felineus was discovered over 100 years ago by K. N. Vinogradov, a prominent pathomorphologist; however, since then the problem of opisthorchiasis has been solved from neither epidemiological nor medical standpoints. Opisthorchiasis remains a widespread disease in West Siberia with an obstinate relapsing course. The opisthorchiasis pattern allowed clinicians to ascribe this disease to systemic human diseases, which require systemic treatment and, correspondingly, systemic and comprehensive knowledge about the liver fluke biology and specific features providing stability of the parasite—host system.
The results of Liver Fluke Genome Project will allow designing highly specific microarray tools for diagnosing opisthorchiasisThe adverse consequences of the liver fluke and other helminthic invasions, which are also of medical and social significance (for example, diphyllobotriosis), should not be underestimated. All these diseases can considerably decrease resistance of the organism, deplete the immune system, complicate accurate diagnostics, and interfere with the efficient treating of other pathologies, thereby considerably decreasing the population labor capacity.
The clinicians of West Siberia have been long concerned with the regional helminthoses. In Novosibirsk, the Institute of Clinical and Experimental Medicine, Siberian Branch of the Russian Academy of Medical Sciences (Prof. A. I. Pal’tsev) and the First Infectious Hospital (Prof. N. P. Tolokonskaya) are involved in the monitoring and detailing of the opisthorchiasis epidemiology, study of the specific features of this pathology, and design of the most efficient treatment strategies. Analogous directions are under development at the Tyumen Institute of Regional Infectious Pathology. However, the bottleneck of all medical studies is the absence of reliable detection methods for opisthorchiasis agent and inability to distinguish it from the other, less abundant, helminthes causing similar symptoms. Neither any treatment schemes nor any prevention measures could be efficient without such diagnostics.
The closed joint-stock company ZAO Vector-Best has made the first step in this direction: the team of T. N Tkachenko has developed a pilot test kit for diagnosing liver fluke invasion based on detection of the patient’s antibodies to liver fluke antigens at various stages of invasion. Unfortunately, this method was found not to be strictly specific for O. felineus. Therefore, the team of V. B. Loktev (Vector-Best) is now developing another test kit for an enzyme immunoassay diagnostics of specific antibodies to a liver fluke protein, paramyosin. First, the scientists cloned the DNA copy of liver fluke paramyosin gene transcript, then produced the pure protein, and used it to obtain the specific antibodies to this protein. This combined genetic engineering and immunological approach is promising for development of specific opisthorchiasis diagnostics.
However, in addition to the improvement of opisthorchiasis diagnostics, the important task in the control of helminthoses is the design of efficient and safe therapeutics. In the developed countries, such drugs are developed based on the state-of-the-art approaches involving genomics, proteomics, metabolomics, bioinformatics, genetic engineering, and combinatorial molecular biology. Such hi-tech approach will be implemented under the Liver Fluke Project at the Siberian Center for Genomics, Proteomics, and Bioinformatics Technologies, which is being established with the Novosibirsk Scientific Center and will be equipped with high throughput equipment for sequencing bacterial and eukaryotic genomes; studying the proteomes (i.e., all proteins) of microorganisms, plants, animals, and humans; and high throughput computations in the field of bioinformatics.
The First Russian Eukaryotic
The new Center is organized under the program of the Siberian Branch of the Russian Academy of Sciences “Genomics, Proteomics, and Bioinformatics”, headed by Academician R. Z. Sagdeev, at the facilities of several institutes of the Branch (Institute of Cytology and Genetics, Institute of Chemical Biology and Fundamental Medicine, International Tomography Center, Institute of Biophysics, and Limnological Institute). For the first time in Russia, the complete cycle of research into bacterial and eukaryotic genomes – starting from their sequencing through computer annotation to prediction of protein spatial structures and reconstruction of gene networks – should be implemented.
The genome of O. felineus is a most appropriate object for a genome project. It is rather compact, amounting to 300 million base pairs, which, according to our studies, are distributed between seven chromosomes. Note that implementation of the first Russian eukaryotic genome project—sequencing of the genome of O. felineus, the agent of opisthorchiasis—became feasible at the Novosibirsk Scientific Center thanks to a unique combination of basic research efforts and practical medicine bolstered up by the state-of-the-art experimental and bioinformatics facilities.
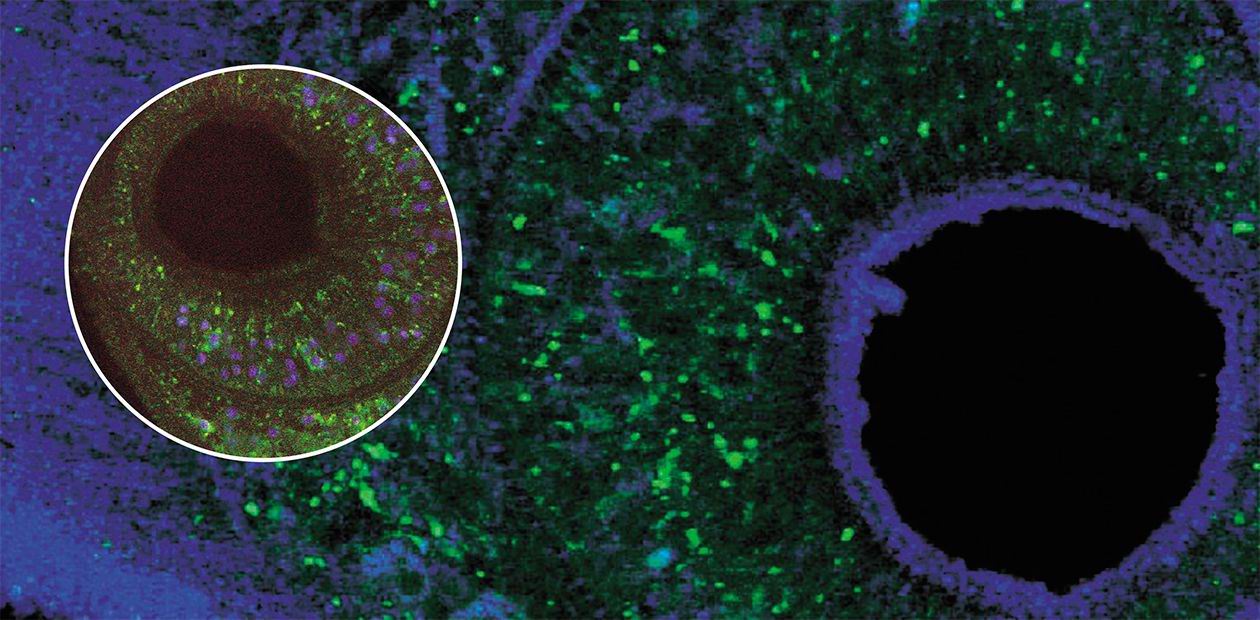
Laser scanning microscopy is able to record individual signals from nanoscale compartments located both on the surface of the analyzed object and deep inside. Successive reading of signals from all “points” in the object makes it possible to reconstruct not only its “optical cross-sections”, but also its spatial structure, providing the insight into fine 3D structure of the liver fluke organs and tissuesThe focus of this project is the development of drugs based on the genomics and proteomics research. Sequencing of the liver fluke genome will determine the molecular targets for specific therapy, i.e., the genes and proteins playing the key role in disease development or control of liver fluke lifecycle and reproduction. These data will be used to model and synthesize the inhibitors blocking the function of target genes. In the long view, the genetic models for opisthorchiasis—experimental animals obtained by genetic engineering, chromosome engineering, and breeding—will also find a wide application.
Creating the future
At the initial stage of the project, the genetic diversity of O. felineus and closely related liver fluke species was for the first time studied in the natural populations of the vast territory comprising the Novosibirsk, Tomsk, and Omsk oblasts and Khanty-Mansi autonomous okrug. These data formed the background for the development of the DNA diagnostics of opisthorchiasis agents, as well as detection of natural opisthorchiasis foci and assessment of the infestation rate.
Activities planned for the second stage include a comprehensive description of the morphology and structure of O. felineus and sequencing its complete genome. For this purpose, the Center is being equipped with the state-of-the-art devices for sequencing, physicochemical analysis, and microscopy.
However, obtaining this information is only half the battle. The current situation in genomics is paradoxical: information volume available to researchers is much larger than they can ponder, analyze, and use in further experiments. Therefore, it is most topical now to develop new mathematical methods, computational techniques, software, and improved methods for description and storage of genomic information. This is the field of bioinformatics, which includes genoinformatics.
The bioinformatics approach has been long and effectively developed at the Institute of Cytology and Genetics; moreover, Siberian bioinformaticians are leaders not only in Russia, but also in the world. Bioinformatics is able to analyze the situation at the four interconnected levels. The first level is genetic text, i.e., the nucleotide sequence of DNA; the second level is also a text but first in an RNA form and then, amino acid sequence of proteins; and the third level is spatial protein structure. And last but not least level is prediction of protein function based on the knowledge of its primary and deduced three-dimensional structures. Thus, structural and comparative genomics expands to a new genomics discipline, which has been named functional genomics.
Functional genomics closely contacts and actually overlaps the new direction in biology named proteomics, whose object is the proteins and their interactions in living organisms, including humans. The Institute of Cytology and Genetics has put into service the most advanced methods of experimental proteomics, such as two-dimensional electrophoresis of proteins and high performance liquid chromatography with subsequent analysis of individual protein fractions with the help of time-of-flight mass spectrometers.
All these methods will be used for analysis and reconstruction of the proteomics portrait of the mature parasitizing O. felineus form. The proteins, peptides, and metabolites secreted by the parasite in interaction with the host organism will be characterized first. This will allow determining the pharmacological targets for rejection of the parasite and restoration of tissues and functions of the damaged organ and subsequent development of highly specific and efficient antiparasitic drugs.
Implementation of such projects in modern biology and biotechnology portends revolutionary changes in pharmacology. Sequencing the genomes of various pathogens will identify constellations of new targets for therapy and this, in its turn, will stimulate the designing of novel drugs, more selective and efficient. Perhaps, the forthcoming pharmacological boom will contribute to a considerable increase in the average human lifespan in highly developed countries, among which we would like to see Russia.
The Liver Fluke Genome Project is the first and so far the only large project of this kind in Russia. We do hope that it will not stand aloof as a Potemkin village in the field of Russian genomics. Thus, we need to identify new problems, generate new ideas, and plan new initiatives, as the tool for implementing such plans—the Siberian Center for Genomics, Proteomics, and Bioinformatics Technologies—will be working at Novosibirsk Akademgorodok many years after the Liver Fluke Genome Project is completed. Possibly, it will become clear with time that this is what this project is about.
References
Beer S.A. The Biology of Opisthorchiasis Agent. — Moscow: KMK Press, 2005, 336 p.
Fedorov K.P., Naumov V.V., Kuznetsova V.G., Belov G.F. Certain real problems in human opisthorchiasis // Med. Parazitol—2002.—V. 3.—Pp. 7—9.
Kaewkes S. Taxonomy and biology of liver flukes // Acta Tropica.—2003.—T. 88.—Pp. 177—186.
King S., Scholz T. Trematodes of the family Opisorchiidae: a mininreview // Korean J. Parasitol.—2001.—V. 201. — Pp. 209—221.
Pauly A., Schuster R., Steuber S. Molecular characterization and differentiation of opistorchiid trematodes of the species Opistorchis felineus (Rivolta, 1884) and Metorchis bilis (Braun, 1790) using polymerase chain reaction // Parasitol. Res. — 2003.—V. 90.—Pp. 409—414.
Wongratanacheewin S., Sermswan R. W., Sirisinha S., Immunology and molecular biology of Opistorchis viverrini infection // Acta Tropica.—2003.—V. 88.—Pp. 95—207.
Contributors to this paper:
N. B. Rubtsov, a doctor of biology, deputy director, head of the Laboratory of Gene Expression Regulation and the Shared Access Center for Microscopic Analysis of Biological Objects with the Institute of Cytology and Genetics Siberian Branch of the Russian Academy of Sciences (ICG SB RAS); S. I. Baiborodin, a candidate of biology, senior researcher with the Laboratory of Gene Expression Regulation (ICG SB RAS); A. V. Katokhin, a candidate of biology, senior researcher with the Sector of Functional Genomics (ICG SB RAS); O. I. Sinitsina, a candidate of biology, senior researcher with the Sector of Mutagenesis and Repair (ICG SB RAS)