The story of the songbird chromosome
This story is a piece of bouts-rimés, a game of rhymed-ends, written with six quills. It tells about the discovery of an “extra” chromosome in the sexual cells of songbirds. Until recently, scientists used to treat this chromosome as a biological oddity since they had found it in two Brazilian species only. Today, Russian geneticists have transformed this exception into a rule for the copious suborder of songbirds. Perhaps, it is this extraordinary genomic feature that creates the tremendous diversity we observe in songbirds and their vocal gifts?
Pavel Borodin: The discovery of the exceptional chromosome
In the beginning of my career path, my teacher, Professor Dmitry Belyaev, gave me a strange task: to look through journals on genetics and write down all cases of strange inheritance of traits — that is, exceptions from Mendel’s laws. At that time, Belyaev was probing the idea of “dormant” genes and was looking for models to check it. And I found quite a few things. Later, we used one of those exceptions — the strange inheritance of the Fused gene in mice — as a model of gene awakening (Belyaev, Ruvinsky, Borodin, 1981). Eventually I described the other exceptions in my book “Essays on Mutants” (1983).
This must have been an imprinting event: ever since, while reading research journals, I’ve been looking for exceptions. Naturally, I could not ignore the story of an extra chromosome in the zebra finch (Taeniopygia guttata), which was discovered in Argentine 21 years ago (Pigozzi, Solari, 1998).

The chromosomal set (karyotype) of cells of the body (somatic cells) of the zebra finch is well-studied. Like many other birds and reptiles, this bird has 40 pairs of chromosomes, with 10 pairs of relatively large macrochromosomes and 30 pairs of very small microchromosomes. Its karyotype is typical for songbirds, or birds in general, for that matter.
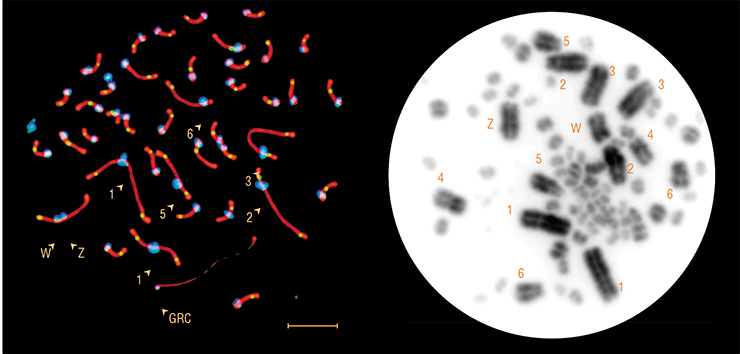
However, the extra chromosome of the zebra finch was not typical at all. It was only present in germ cells, where it was the largest macrochromosome. In somatic cells, it was absent. That is why Pigozzi and Solari called it germline-restricted chromosome (GRC).
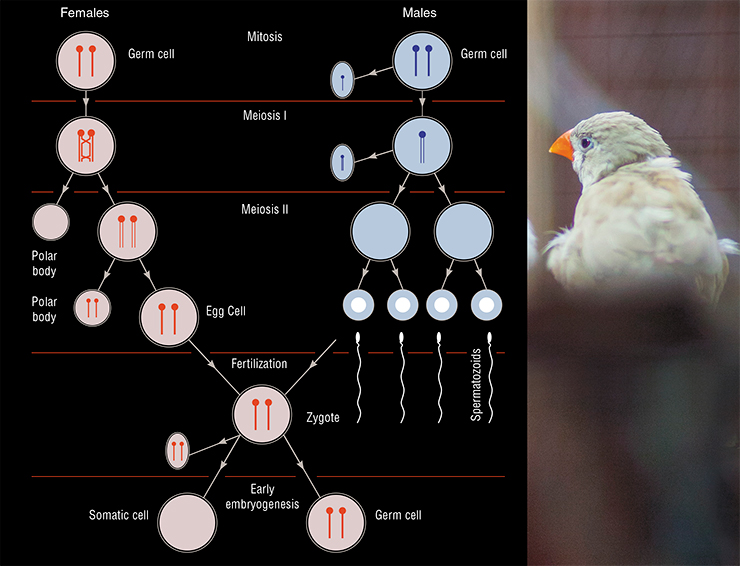
Oocytes, usually contained two copies of the GRC, or, more rarely, a single copy. In females, paired GRC’s usually behaved almost like regular chromosomes. In the beginning of meiosis (cell division, which results in halved chromosome number), they found each other, conjugated along their whole length and recombined, exchanging their segments with each other. For some unknown reason, these exchanges almost always occurred at the chromosomes’ ends, while in other chromosomes, exchanges could happen in all regions. One of the females in Pigozzi and Solari’s study had just one GRC, which did not conjugate or recombine with anything.
All males carried a single GRC in their spermatocytes (immature sperm cells), and only did so until the first meiotic division: at that stage, or shortly after, it was expelled from the cell. Spermatocyte preparations were speckled with tiny round dense granules — the ejected GRC’s. As the result, mature sperm cells did not have those chromosomes. And yet, all germ cells, oocytes and immature spermatocytes had at least a single copy of the GRC. This means they got it from females. But why did males always have a single GRC, while females usually had two?
Pigozzi and Solari came up with a complex scheme of GRC inheritance, where it is passed down to offspring as a single copy via females (Pigozzi, Solari, 2005). According to this scheme, it is present in the zygote and embryonic stem cells, but eventually all cells, except gamete progenitor cells, lose it. To explain the origin of the second copy of the GRC in female oocytes, Pigozzi and Solari suggested that it duplicates in oocytes, but not in spermatocytes.
The scientific community put this remarkable chromosome into the long list of biological exceptions and carried on. Five years ago, Maria Pigozzi found a similar chromosome in the Bengalese finch (Lonchura striata domestica) (del Priore, Pigozzi, 2014). But this did not make the GRC any less exceptional, because the two species are closely related.
Anna Torgasheva: the first swallow
The GRC of the zebra finch remained an exception until June 25, 2015, when I found it in the sand martin (Riparia riparia). I was not looking for that chromosome. I was working on my part of a project on the analysis of frequency and distribution of recombination in avian genomes. Together with Elena Kizilova and Marina Rodionova from our institute, we were extracting gametes from bird ovaries at early stages of meiosis and stained the chromosomes using antibodies to meiotic chromosome proteins.
Chromosomes of all birds I had worked with before looked absolutely normal. This chromosome of female sand martin looked completely aberrant. It was bigger than all other chromosomes, and it had no pair. At first, I thought it was a sex chromosome — in some birds, they occur in very intricate configurations. But this martin had a completely normal ZW pair. And it had this abnormal chromosome.
When I showed the photo to Borodin., he screamed “It’s the GRC! It’s the GRC!” and handed me the zebra finch paper. Our chromosome looked identical to the zebra finch GRC. It was one of the largest chromosomes, with a single copy in males and (usually) with two copies in females — although the very first female had a single GRC, which was exactly why I noticed it. I asked Ekaterina Basheva to make chromosomal preparations from sand martin bone marrow. These preparations did not have the odd chromosome. Inna Pristyazhnyuk made preparations from cultured fibroblasts — and it was not there, either. The chromosome was only present in germ cells. Indeed, this was the GRC. In a sand martin!
Lyubov’ Malinovskaya: The private life of the GRC
Shortly after the discovery of the GRC in martins, I came to Borodin’s lab, and they entrusted me with an in-depth study of meiosis in birds. At the first glance, the sand martin GRC behaved in the very same way as the zebra finch GRC. In females with two GRC copies, we observed their complete conjugation during the first stage of meiosis, and recombination points were located in the same regions as in the zebra finch — at both ends of the GRC.
The proportion of single-copy GRC females was the same as in zebra finches (13 %), even though our sand martins came from different natural populations, and zebra finches came from a pet shop. In all these females, their solitary GRC did not attempt to conjugate or recombine with any chromosomes from the main set.
This is why I did not expect to see anything new when I began the analysis of male germ cells. In spread nucleus preparations of spermatocytes, the GRC is often present as a tangled strand, as opposed to even and straight regular chromosomes. Sometimes I found tangled lumps twice the regular size, but attributed it to the specifics of preparation method. The intensity of GRC staining at different preparations may vary a lot..
However, as I saw more and more of these cells, doubt creeped in. At one moment, I clearly saw two centromere signals, which mark regions holding sister chromatids together. I could not believe my eyes, adjusted the image contrast and saw two distinct individual strands, clearly indicating that there were two GRC copies in a single cell!
Instead of rushing to announce it right away, I thoroughly checked all previous images and ended up with a few dozen cells. It was beyond doubt — what I was seeing was neither an artefact of the method nor a random transfer of GRC from an adjacent cell. Almost all males were mosaic by the number or the GRC copies. The share of double-GRC spermatocytes ranged between 4 and 60 percent.

Where do these mosaics come from? According to the model proposed by Pigozzi and Solari, both males and females inherit a single copy of GRC from their mothers. It duplicates in all gamete progenitor cells of females and only in some — in males. We believe that males and females inherit either one or two copies of GRC from their mothers. Females with two copies keep them until the end and pass them on to their progeny. Males with two copies often lose one of them in the regular (mitotic) cell divisions, and lose the other one during the first meiotic division.
In any case, our data suggest that both in sand martins and zebra finches, all germ cells carry at least one copy of the GRC. In males, it persists until the middle of meiosis, and until the end — in females. We have not found a single egoocyte without the GRC.
Anna Torgasheva: GRC in other species
 If sand martin GRC looks and behaves exactly in the same way as that of the zebra finch, does that mean that it was present in the common ancestor of the two species, which lived 35 million years ago (Kumar et al., 2017)? Has it been inherited, virtually unchanged, by all of its progeny? In this case, many other songbirds must have it, too — and there are about 5 thousand of them. Why has it never been seen in any other species except two finches and a martin? The answer is simple — no one has ever bothered to look for it. No one has ever analyzed the germ cells of these birds.
If sand martin GRC looks and behaves exactly in the same way as that of the zebra finch, does that mean that it was present in the common ancestor of the two species, which lived 35 million years ago (Kumar et al., 2017)? Has it been inherited, virtually unchanged, by all of its progeny? In this case, many other songbirds must have it, too — and there are about 5 thousand of them. Why has it never been seen in any other species except two finches and a martin? The answer is simple — no one has ever bothered to look for it. No one has ever analyzed the germ cells of these birds.
We realized that this was precisely what we had to do, and began our search from the closest relative of the sand martin. We were not too surprised to find exactly the same GRC in the pale martin (Riparia diluta). The two species diverged relatively recently — around 2 million years ago.
The next accessible species was the barn swallow (Hirundo rustica). The accessibility is a relative concept — to get my hands on it, I had to climb a rickety ladder to reach the roof of a high-ceilinged garage; Borodin. was securing the ladder with one hand, while keeping the other hand on his racing heart. At one moment, he even let go of the ladder to take a picture.
Unfortunately, our effort was fruitless — not under the ceiling, but under the microscope. Female barn swallows had no extra or odd macrochromosomes. This was a strong argument against the common origin of the GRC in the two other martin species, on the one hand, and two finch species, on the other hand. However, there was a possibility that the barn swallow itself was an exception and had lost its GRC in the course of its 18 million year evolutionary independence from martins. We had to continue our search in the neighboring branches of the phylogenetic tree.
We acquired most of the birds for the analysis from Elena Shnaider from the Sibekotsentr (Siberian Ecological Center). They were dead or lethally wounded birds that were brought to their Raptor Rehabilitation Center). In the spring of 2016, Elena brought us a dead male bullfinch. We discovered the GRC in its germ cells. Again, it was the largest chromosome and had no pair. Again, it was exactly the same as in martins and finches. But the bullfinch belongs to a completely different branch of the avian phylogeny and is almost equally distant from both finches and martins.
Apparently, it was too early to discard the common ancestry hypothesis. So we began to systematically check it. Actually, the check was not that systematic — we just analyzed chromosomes of all birds we could lay our hands on. In the majority of songbirds, we found large GRC that was absent in their somatic cells.
And yet, some of these species did not have any macrochromosomes in their gametes. They were distributed across the phylogenetic tree without any apparent logic. We did not find the GRC in the barn swallow, rook, goldfinch, canary, pine bunting, or flycatcher. That could mean that the GRC emerged once in the common ancestor of all songbirds, but some species later lost it in the course of their independent evolution, and that the GRC is not that important and indispensable.
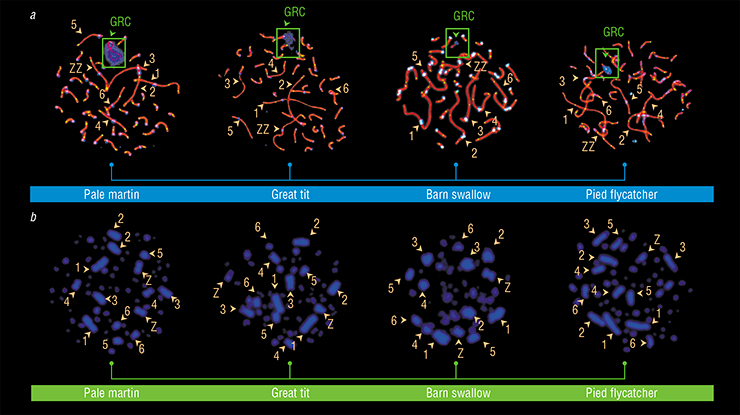
I was trying to figure out what those GRC-less species had in common, when it occurred to me that we’d been analyzing only female chromosomes — and in females, the GRC behaves like a regular chromosome pair during meiosis — it conjugates and recombines… Which means that if it were not the unambiguously largest chromosome, it would be impossible to distinguish it from other paired chromosomes. However, in males it would be apparent due to its oddity, the lack of recombination signals and the characteristic diffused cloud of chromatin, which, for a reason yet unknown, binds antibodies against centromeres.
We launched a hectic search for a male barn swallow. We managed to catch the bird in an abandoned brick shed. And… we saw a tiny odd chromosome it its spermatocytes. Non-recombining and surrounded by a swarm of antibodies. It was the GRC.
We found the same tiny chromosome in flycatchers, goldfinches, canaries, pine bunting, and rooks. We found the GRC in all of the 14 songbird species that we studied, and the two previously studied species of finches.
To sum up: the GRC is present in 16 songbirds from 9 different families. It is probably present in all other 5000 songbird species. Songbirds are a clade in the order Passeriformes. This order also contains the suboscines (Tyranni) with about a thousand species with a predominantly South American distribution. The two clades split about 50 million years ago. So far we have been unable to acquire any suboscines and we do not know if they have the GRC. What do we know about other birds? Perhaps they have the GRC, too, but it simply has not been found yet?
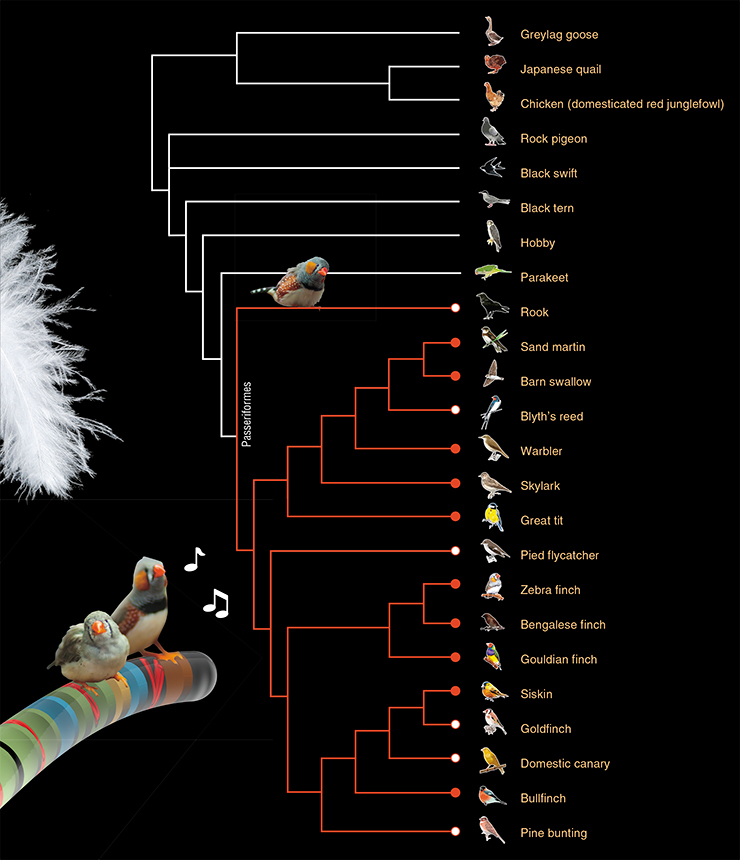
We have scrutinized all available photographs of meiotic chromosomes in male birds from different orders. There is definitely no GRC in chickens, quails, and pigeons. These three birds have been examined by Maria Pigozzi, and she would have seen the GRC if it had been there. We examined spermatocytes of geese, terns, swifts, hobby falcons and parakeets (parrots are thought to be the closest relatives of passerine birds). None of them had anything resembling the GRC.
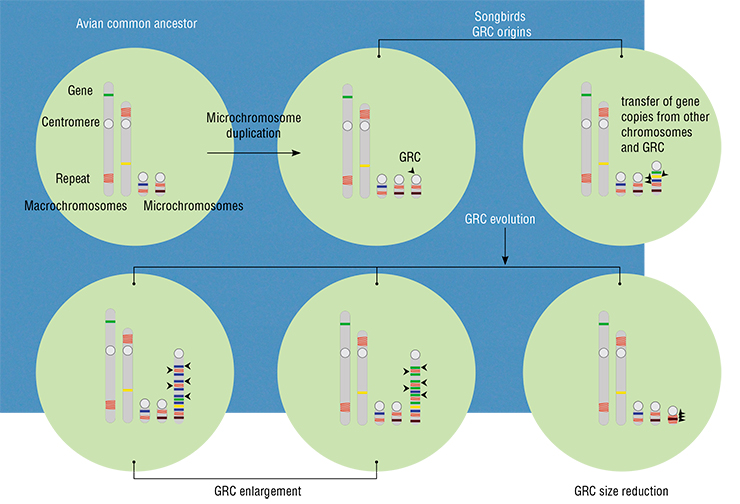
So, none of the ten non-passerine species had the GRC. It must have appeared once in the common ancestor of all songbirds, i. e. around 35 million years ago, or about 50 million years ago in the common ancestor of all passerines (Kumar et al., 2017). What has it been through all this time? We know that it has been changing in shape and size. But what about its content?

To answer this, we needed a molecular probe to compare the genetic content of the GRC in different species. We found it across the corridor, in the lab of Nikolai Rubtsov, an expert on chromosome microsurgery.
Nikolai Rubtsov: the GRC Probe
In my research of the organization and evolution of chromosomes, I have always looked for exceptions. In humans, it was small supernumerary marker chromosomes; in other eukaryotes — extra B-chromosomes, and various signs of chromosomal instability in general. When Borodin asked me to join the search for the nature of the GRC, I was thrilled. Immediately, I began to study the discovered GRC with the techniques developed in our laboratory such aschromosome microdissection and creation of DNA-libraries of individual chromosomes and chromosome regions
Collecting GRC material turned out to be relatively easy. Normally, we must enter the nucleus, choose the necessary chromosome or a specific region — without accidentally grabbing the adjacent chromosomes. With the GRC, I was looking for cells where it had been ejected from the nucleus and was present as a round dense body lying nearby. Using an electronic micromanipulator, I moved the GRC to an open space with a needle and, using a microscope, picked it with a pipette with drop of special solution in its disposable tip and transferred it into a tube.
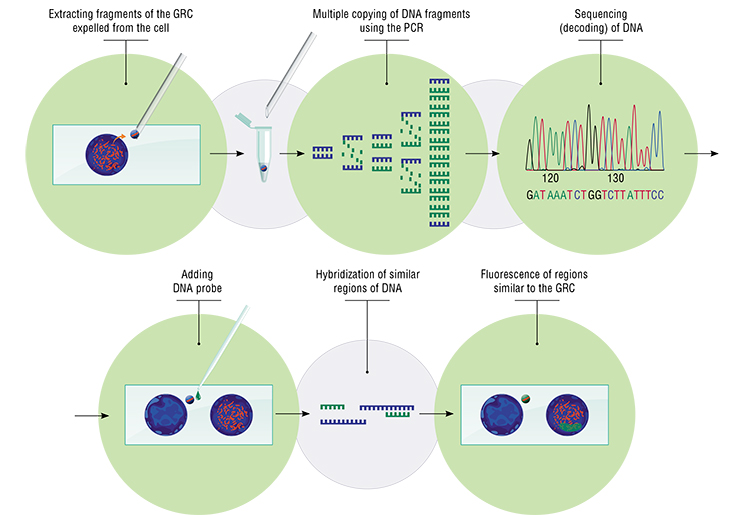
My colleagues Kira Zadesenets and Tatiana Karamysheva extracted DNA from this material and amplified it (i. e. made multiple copies) using the polymerase chain reaction (PCR). We used the resulting material to sequence and analyze the genetic content of the GRC. We used the same material to create a DNA probe — a set of small DNA fragments from this chromosome up to 400 base pairs long, in millions of copies. These fragments were marked with fluorochromes (dyes with luminescent properties).
We now had GRC probes for four species of birds — zebra finch, Bengalese finch, pale martin, and siskin (Carduelis spinus) — which could be applied to chromosome preparations of different species. After denaturation and renaturation, marked fragments of the GRC were supposed to hybridize (bond) with homologous (i. e. similar and related) regions in the sample specimen. These regions can be seen in a luminescent microscope.
The probes were designed to detect the GRC in any condition, in any cell, and at any stage of the cell cycle, and to analyze its genetic contents. With these probes, we could compare the GRC’s of different species, and search for homologies between the GRC and regions of the main genome.
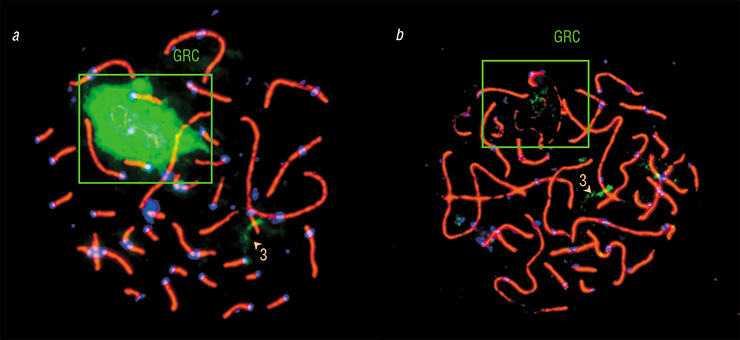
Just as expected, DNA from the expelled GRC hybridized with the GRC inside germ cells. Moreover, we observed clear signals of hybridization with certain regions of chromosomes from the main set. This meant that the GRC contained copies of this DNA. Interestingly, in different species, the GRC contained copies of different genomic regions.
Overall, the GRC of different species varied substantially in terms of its genetic content. For instance, zebra finch GRC DNA probe hybridized with only about a half of the Bengalese finch th e GRC and virtually did not bind with martin GRC. On the other hand, large GRC DNA probe of the pale martin marked small GRC of the barn swallow, indicating their common origin. This was very uplifting news — at last, we had molecular proof that the small GRC was in fact a GRC.
Svetlana Galkina: “Lamp brushes” made of GRC
In the summer of 2015 I was in my lab the Chair of Genetics of the St. Petersburg State University, when my phone rung. It was Borodin .: “We found the GRC in sand martins. Don’t tell anyone yet! It’s the same as in the zebra finch. Rubtsov made us GRC probes for sand martins and zebra finches. It’d be great to know what it looks like in growing oocytes and what it does there”.
I have known about the extra chromosome of the zebra finch for quite a while, because my scientific interests include the arrangement of chromosomes in growing avian oocytes. In birds, the oocyte accumulates yolk and grows to a giant size — that is, it becomes an egg. As it begins to mature, great amounts of RNA are synthesized on its chromosomes. These nucleic acid molecules serve as matrix to produce proteins or perform regulatory functions.
During this process, chromosomes begin to look like bottle brushes — or, more specifically, like brushes used in the XIX century to clean kerosene lamps. This was what they looked like to their discoverers; they have been called lampbrush chromosomes since. At this stage, they reach gigantic sizes (on the cellular level) — up to one or two tenths of a millimeter. They can be manipulated manually under a low-magnification microscope. The technique of making lampbrush chromosome preparations is not complicated, but it requires sharp eyes and skilled hands to scan the yolk surface and find the translucent nucleus, surrounded only by the slightly opalescent nuclear envelope.
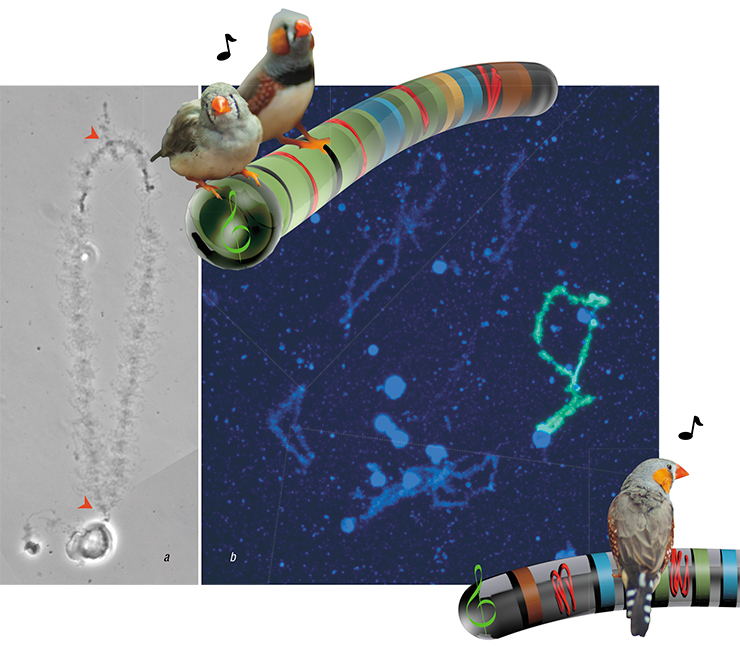
We had our first lampbrush preparations from zebra finch oocytes in 2010. We saw the largest chromosome and assumed it was the GRC. Of course, we expected the GRC at this stage to stand out visually — after all, it was so special! Alas, it turned out that during the lamp brush stage, this biggest pair of chromosomes looked exactly like regular chromosomes. How were we supposed to prove it was the GRC? We took pictures of the preparations of the “unproven” chromosomes and put them aside — for the next six years. We needed a specific probe for that chromosome, and back then, we did not have it.
When Rubtsov sent us the probe, we — Alsu Saifitdinova, Lera Volod’kina and I — made new preparations and hybridized it on a zebra finch preparation. We got direct proof that the suspicious chromosome was indeed the GRC at the lampbrush stage, and that it behaved just like any other chromosome at that stage — it was actively reading information from DNA to RNA. But what was this information?
Anna Torgasheva: What is GRC made of
We faced the main question: what was the GRC made of? What genes did it contain? What else was there, apart from genes? How similar or different are the GRC’s of different species? To answer these questions, we sequenced the DNA that Rubtsov got from the ejected GRCs of the zebra and Bengalese finches, siskin, and pale martin.
Unfortunately, this method has its limitations and not all regions of the DNA get analyzed. This means it is impossible to obtain the complete sequence. Moreover, as we have learned from the results of cross-hybridization, the GRC contains many repeats. This complicated the task even further.
Therefore, we began to compare the obtained sequences with known sequences of the zebra finch genome. Of course, this way we could not establish whether the GRC contained any unique genes, but we could see what it had in common with the DNA of the main set of chromosomes.
It turned out that in each of the species, the GRC contained copies of short (up to 2500 base pairs long) regions of the main genome, repeated tens to several thousand times. They were different in all species. It was very similar to what we saw in the hybridization experiments. It seemed likely that in different bird lineages, different sequences had been copied to and multiplied in the GRC.
What did these regions contain? Unfortunately, nothing special. They were mostly noncoding regions of genes (introns) and intergenic regions. However, there were some coding regions, too, but they were overall rare and incomplete.
Denis Larkin: In the light of avian genome evolution
To say that the avian genome is “depressingly conservative” is like calling the Sun boring. There may be billions of other stars around, and many of them are way more spectacular than our Sun, but they do not keep us warm. It is the same with the avian genome. Indeed, compared to the insanely mobile mammalian genomes, the avian genome is very conservative and steady-going. On the other hand, if your genome is so efficient that it makes you fly (like, literally fly), why change it at all? You could fall!
To learn to fly, birds had to sacrifice the majority of their mobile genetic elements, which are the drivers of evolution in animals (Kazazian, 2004). In mammals, these mobile elements do all sorts of things: form new genes, change regulation in existing genes, and reconfigure chromosomes — all that for the greater purpose of survival and adaptation in the constantly changing environment. Keep up — or… tough luck!
To a detached observer, the avian genome may seem like a boring place compared to mammalian genomes. This is not true. The evolutionary process never stops, because a stop means instant (which, in evolution, stands for about a million years) extinction. What the Black Queen said to Alice — «Now, here, you see, it takes all the running you can do, to keep in the same place. If you want to get somewhere else, you must run at least twice as fast as that!»– fully applies to evolution. You need to change all the time to appear unchanged in the ever-changing world. To actually change, you need to do it even faster.
This produces a sort of an avian paradox: to fly, it is better not to change; but to stay on the face of Earth, you need to change. Birds have learned the lesson of their seniors — the dinosaurs. To survive in unfavorable conditions, you need to be small, and preferably be able to flee (fly) from serious difficulties instead of trying to overcome (adapt to) them. However, this holds a risk of losing to other animals instead of the general environment — the harsh rules of evolution state that two species cannot coexist in the same ecological niche. One of them will have to give in.
If we look at birds as a whole, almost a half of all ten thousand species are songbirds. To link this amazing diversity to the GRC is very tempting.
Together with my colleagues, we have established quite a while ago that in mammals, regions of chromosomes where their crazy genomes split to form new structures (and new species) carry genes that are explicitly connected with the most outstanding features of these species. For instance, in ruminants, it is the genes called defensins, which protect them from toxins produced by myriads of microbes inhabiting their rumen. In pigs, it is genes responsible for their sense of taste, allowing these animals to consume even the least attractive of foods.
We now know that regions of such chromosomal rearrangements in mammals accumulate mobile genetic elements that change the expression of the necessary genes (this is completely random, but natural selection never sleeps). This does not happen in species with no such rearrangements.
Birds have few mobile elements, and changing something is a challenge. When P. M. told me that he had “decoded” fragments of this unusual chromosome, my first thought was — we should check if the GRC was a sort of a junkyard for chromosome “shreds” that form during chromosome rearrangements. In this case, they had to look like DNA from rearranged regions in their ancestral genomes or like DNA from similar but non-arranged genome regions of other birds. It turned out that there was no similarity — at least not in the zebra finch, and we did not have any other genomes.
We did, however, learn a peculiar thing: even closely related species have very different GRC sequences. Moreover, the GRC has many mobile elements that were active in the common ancestor of songbirds, but very few mobile elements that were active in the common ancestor of all birds. This allowed us to date the origin of this chromosome in yet another manner. Apparently, it was absent in the common ancestor of all birds, and the few ancient mobile elements were probably transferred to the GRC from the somatic genome together with other gene sequences. But in the common ancestor of songbirds, it was already there.
This does not answer the question of the influence of the GRC on avian speciation. The great diversity of songbirds is a telling sign that songbirds have found an escape from the dullness of the basic genome without really changing the way it works. Recently, we discovered that roughly at the same time with the emergence of the GRC, songbirds began to experience major rearrangementsof the gene-rich small chromosomes. Could it have been the influence of the GRC with its “duplicate” chunks of the genome? This remains to be found out… or not.
Pavel Borodin: How were we writing our GRC paper
We discovered the “first swallow” in June, 2015; by 2018, we had all the material ready — it was time to put it into a good paper. Actually, we began writing back in the fall of 2016, when we only had two martins — the sand martin and the pale martin, and the first results of cross-hybridization of DNA between them and the finches. Back then, we firmly believed in recent and independent origin of the GRC in these two phylogenetic lineages. As new data kept pouring in, we took our time to rewrite the paper.
All these years, the first thing I did in the morning was google “GRC songbirds”. I had a nagging premonition that someone would outrun us and publish a paper titled “Did you know that not only zebra finches have the GRC…”.

On May 3, 2018, when we were on version #33 of our draft, Current Biology published a paper by American researchers titled Discovery of the First Germline-Restricted Gene by Subtractive Transcriptomic Analysis in the Zebra Finch, Taeniopygia guttata. The authors discovered a gene called aSNAP in the zebra finch GRC, and in the main genome, they found the original copy of this gene, which codes a protein necessary in the process of plasmatic membrane fusion.
No one knows what this gene does in the GRC, but it turned out to be substantially different from the original. This means that it got into the GRC a long time ago. The last phrase in the paper said: “our data imply that the GRC is relatively old and may be present in more bird lineages than originally expected.” In a very similar situation, Darwin wrote that Wallace’s paper “contained exactly the same theory as mine ”.
Without consulting Lyell and Hooker, we dropped everything and in two months finally finished with our paper. Being honest people, we sent it to the same journal that had published our “Wallace letter”, that is, Current Biology. But they did not even bother to send it to reviewers and returned it two days later, suggesting that we send it to a more specialized journal — they must have decided that for a general biology journal like theirs, one GRC paper would suffice for years to come.
Naturally, we did not send our discovery to a specialized journal. No way! It definitely deserved the attention of the whole humankind. Our main secret was out in the very title of the paper — Germline-restricted chromosome (GRC) is widespread among songbirds. Immediately, we uploaded the paper to a preprint website called bioRχiv.org. After that, we made a list of journals fit to publish our discovery and began mailing their editors.
While we were collecting rejections rejections, the same website, bioRχiv.org, published a preprint of a joint Swedish-Dutch-American-German-Spanish team titled Programmed DNA elimination of germline development genes in songbirds (Kinsella et al., 2018).
The authors of this paper have found that
• at the lowest estimate, zebra finch GRC carries over a hundred genes, with one third active primarily in the ovaries and only six — in the testicles. The products of these genes take part in the development of the reproductive system, particularly female, as well as in various processes of cellular development, including neuron differentiation.
• The original copies of all genes found in the GRC were present in the main genome of the zebra finch and other birds.
• Among the GRC genes of the zebra finch, there are two which were probably copied to the GRC a very long time ago, way before the irradiation that produced the current diversity of songbirds, and has changed significantly since.
The latter implies that the “the GRC is tens of millions of years old and likely present across songbirds (…), consistent with [the] recent study”. They were referring to our prerprint, which was an honest and noble thing to do.
What amazes me the most in this story that three independent research teams came to the same conclusion simultaneously, from different points of view and based on different material: the GRC is widespread among songbirds. This is, verbatim, the title of the paper that was printed in July 2019, i. e. precisely four years after the “first martin”, by one of the world’s leading interdisciplinary journals — Proceedings of the National Academy of Sciences (a.k.a. PNAS) (Torgasheva et al., 2019). The paper was featured on the cover of the issue.
When the paper came out, we were basking in fame. New Scientist, GenomeWeb, N+1 as well as TASS and Rossiyskaya Gazeta (both are major government media), Cherdak, gazeta.ru and even Pikabu (“the Russian Reddit”) all wrote about us. But a poet was right: fame is but a gaudy patch on the bard’s tattered rags. We learned a lot about our work from the press: for one, songbirds can sing only because of their special chromosome — they would be mute otherwise. Also, bird females owe their femaleness to the extra chromosome in their eggs.
Pavel Borodin and Anna Torgasheva: Homework for tomorrow
And finally, a short list of GRC-related questions.
First, when, why and how is it expelled from somatic cells?
 • When: how early in the course of ontogenesis does it happen? The science knows several cases of whole chromosomes or their regions expelled from somatic cells. This phenomenon, called chromatin diminution and programmed DNA elimination, has been known for over a century. It has been described for many animals: roundworms, cyclops, fungus gnats, lampreys, and hagfish. In the cases when the process was studied, it was found to occur very early in the development, usually during the very first divisions of the zygote. We do not know anything about birds yet.
• When: how early in the course of ontogenesis does it happen? The science knows several cases of whole chromosomes or their regions expelled from somatic cells. This phenomenon, called chromatin diminution and programmed DNA elimination, has been known for over a century. It has been described for many animals: roundworms, cyclops, fungus gnats, lampreys, and hagfish. In the cases when the process was studied, it was found to occur very early in the development, usually during the very first divisions of the zygote. We do not know anything about birds yet.
• Why (and what is the adaptive value): to decrease the somatic cell genome size and its maintenance costs? To rid somatic cells of genes necessary for early development and useless or even detrimental in a mature organism? Could the GRC be a sort of a setup file?
• How: in the course of cell divisions, the GRC passively lags behind compared to other chromosomes. Is it because it has a flimsy centromere, and the spindle apparatus has a hard time sticking to it? Or is it because cells actively kick it out from their nucleus?
Second, when did it appear? We know that all songbirds have the GRC, and none of the birds outside the Passeriformes have it. What we do not know yet is if the Tyranni have it. These birds inhabit Australia, South America and South-East Asia. There are no tyrannid residents or vagrants in Siberia, and you cannot find them in pet shops. But if they turn out to lack the GRC, it would mean that it appeared in the common ancestor of all songbirds about 35 million years ago. If the Tyranni have it, it points at the common ancestor of all passerine birds and makes the GRC 20 million years older.
Third: perhaps it was this exceptional chromosome that is at the root of the mind-blowing diversity of passerine birds and rapid evolution of their amazing and beautiful forms and songs? If it is true, how does it work?
Afterword
Treasure your exceptions!... Exceptions are like the rough brickwork of a growing building which tells that there is more to come and shows where the next construction is to be.
William Bateson
References
Borodin P. M. Etudes about mutants. М.: Znanie, 1983. 112 p. [in Russian].
Bateson W. The Method and Scope of Genetics. 1908. 45 p.
Belyaev D., Ruvinsky A. & Borodin P. Inheritance of alternative states of the fused gene in mice // Journal of Heredity. 1981. V. 72. P. 107–112.
Biederman M. K., Nelson M. M., Asalone K. C. et al. Discovery of the First Germline-Restricted Gene by Subtractive Transcriptomic Analysis in the Zebra Finch, Taeniopygia guttata // Curr Biol. 2018. V. 28. N. 10. P. 1620–1627.
del Priore L. & Pigozzi M. I. Histone modifications related to chromosome silencing and elimination during male meiosis in Bengalese finch // Chromosoma. 2014. V. 123. V. 3. P. 293–302.
Itoh Y. & Arnold A. P. Chromosomal polymorphism and comparative painting analysis in the zebra finch // Chromosom Res. 2005. V. 13. N. 1. P. 47–56.
Kazazian H. H. Jr. Mobile elements: drivers of genome evolution // Science. 2004. V. 303. N. 5664. P. 1626–1632.
Kinsella C. M., Ruiz-Ruano F. J., Dion-Côté A-M. et al. Programmed DNA elimination of germline development genes in songbirds // bioRxiv. 2018. doi:10.1101/444364.
Kumar S., Stecher G., Suleski M. & Hedges S. B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times // Mol Biol Evol. 2017. V. 34. N. 7. P. 1812–1819.
Pigozzi M. I. & Solari A. J. Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, Taeniopygia guttata // Chromosome Res. 1998. V. 6. N. 2. P. 105–113.
Pigozzi M. I. & Solari A. J. The germ-line-restricted chromosome in the zebra finch: recombination in females and elimination in males // Chromosoma. 2005. V. 114. N. 6. P. 403–409.
Torgasheva A. A., Malinovskaya L. P., Zadesenets K. S. et al. Germline-restricted chromosome (the GRC) is widespread among songbirds // Proceedings of the National Academy of Sciences. 2019. V. 116. N. 24. P. 11845–11850.
The zebra finch photos in this article are courtesy of M. Kuleshin; photos of other birds – courtesy of N. Andreenkova, O. Andreenkov, and E. Shnaider