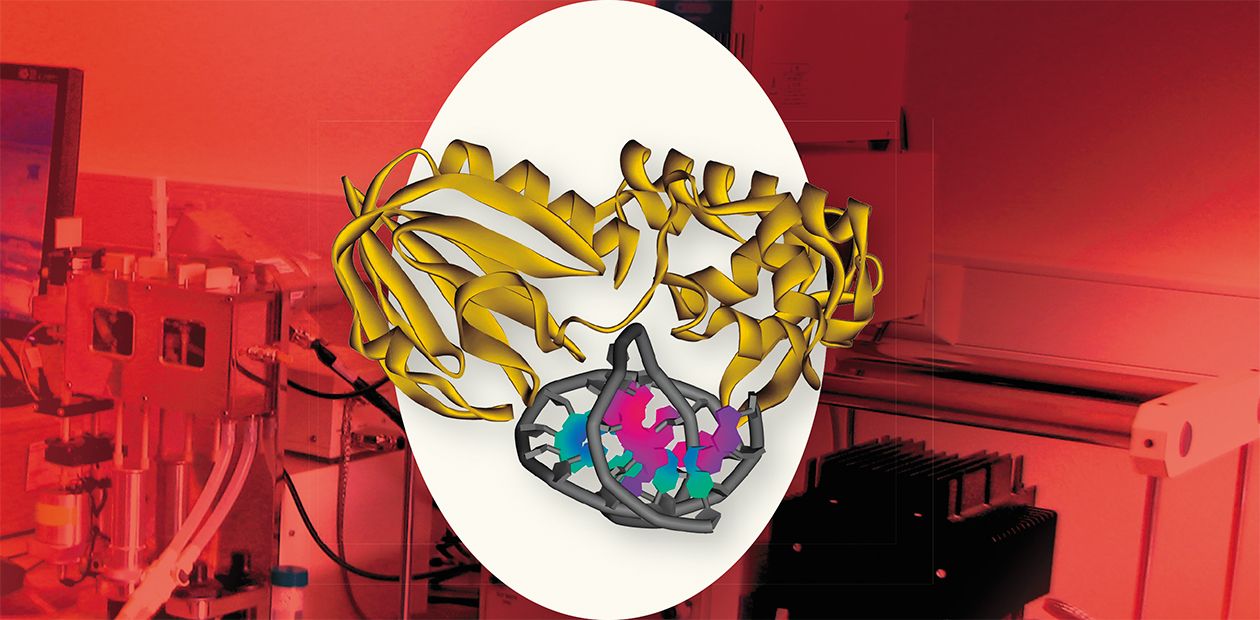
Enzyme Starring
Biochemistry of any living organism can be regarded as a set of enzymatic reactions, which is not much of a hyperbole. Unlike common catalysts, enzymes, being catalysts of a protein nature, display an extremely high efficiency and specificity. Moreover, both the enzyme and the substance undergoing the chemical transformation are large and complex macromolecules with variable spatial structures. The insight into the detailed mechanism underlying the interaction of an enzyme with its substrate is of keen theoretical and practical interest, but it is a difficult and nontrivial problem requiring a wide range of state-of-the-art research tools
Biochemistry of any living organism can be regarded as a set of enzymatic reactions, which is not much of a hyperbole. Unlike common catalysts, enzymes, being catalysts of a protein nature, display an extremely high efficiency and specificity. Moreover, both the enzyme and the substance undergoing the chemical transformation are large and complex macromolecules with variable spatial structures. The insight into the detailed mechanism underlying the interaction of an enzyme with its substrate is of keen theoretical and practical interest, but it is a difficult and nontrivial problem requiring a wide range of state-of-the-art research tools
How do the molecules of an enzyme and its substrate interact in an enzymatic reaction? It is by no means simple to understand all the details of this vitally important process. Actually, researchers may have at their disposal a crystal of either a free enzyme or its complex with the final reaction product. It is practically impossible to obtain such a crystal at a certain intermediate stage of the enzymatic reaction (for example, directly at the stage of catalysis).
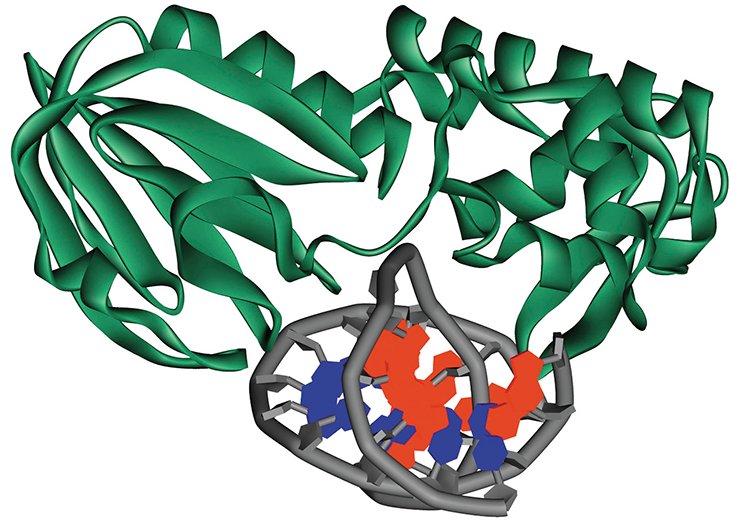
Indeed, modern molecular biological methods can help to deal with this situation. For example, it is possible to modify the substrate so that it would halt catalysis but would not influence the substrate binding to the enzyme; as a result, we obtain the crystal of the corresponding intermediate complex. However, this approach allows us to study only a limited number of enzyme states. As for the “crystal-free” approaches to determining the structure of molecules, such as nuclear magnetic resonance (NMR), their application is rather limited because of the difficulties in data interpretation.
The structural data obtained by the above mentioned methods are in fact “snapshots” of particular structural rearrangements in the molecules of an enzyme and substrate. However, this is insufficient for the current level of scientific knowledge, which demands a detailed full-length “movie.”
DNA-Guarding Forces
It is well-known that dozens of enzymes in the living cell are constantly safeguarding genetic information from damages. This is necessary because the cellular DNA, while functioning, is subject to negative impacts of various physical and chemical environmental factors such as ultraviolet and radioactive radiation, carcinogenic chemicals, and others. Among the most aggressive factors are reactive oxygen species, which cause the so-called oxidative DNA damages. Such damages in the genetic machinery display cytotoxic and mutagenic effects and thereby can induce various cardiovascular, neurodegenerative, and cancer diseases.
The cell “mends” its DNA with the help of a specialized. The enzymes of this system can rapidly and precisely locate the damages among a tremendous number of the nucleotide units constituting a DNA strand and initiate their repair. While searching for the damaged nucleotides, the molecules of repair enzymes can “jump” from one DNA region to another between different DNA regions within the cell nucleus or just successively crawl along the DNA strand, each of these enzymes being able to specifically recognize its own damage.
Research into the mechanisms and specific features of the operation of DNA repair enzymes has recently aroused a special interest, since this kind of knowledge of can help the mankind to solve the problems of early aging and of controlling over the diseases caused by genetic mutations.
A fluorescent video
It is also possible to record the changes in enzyme and DNA structures directly during their interaction with the help of optical methods.
As is known, tryptophan is the most intensively fluorescing amino acid, accounting for about 90% of the overall intrinsic protein fluorescence. Therefore, tryptophan is used as a highly sensitive marker when studying conformational changes in protein molecules, enzymes included. However, when the intrinsic fluorescence of macromolecules is insufficient for obtaining the information of interest, it is possible to use special substances, fluorophores, which display the required photophysical properties.
It is possible to make a real “video recording” of enzymatic process using instead of a video camera the so-called stopped-flow spectrometers, which can record rapid changes of the fluorescence intensity in a solution.
This method was used in the Laboratory of Biopolymers Modifications, Institute of Chemical Biology and Fundamental Medicine, Siberian Branch, Russian Academy of Sciences (Novosibirsk, Russia) to study the dynamics of changes in the spatial structure of one of the DNA repair enzymes, formamidopyrimidine-DNA glycosylase (Fpg) from the bacterium Escherichia coli, during its interaction with several DNA species containing different damaged nucleotides (Koval et al., 2010).
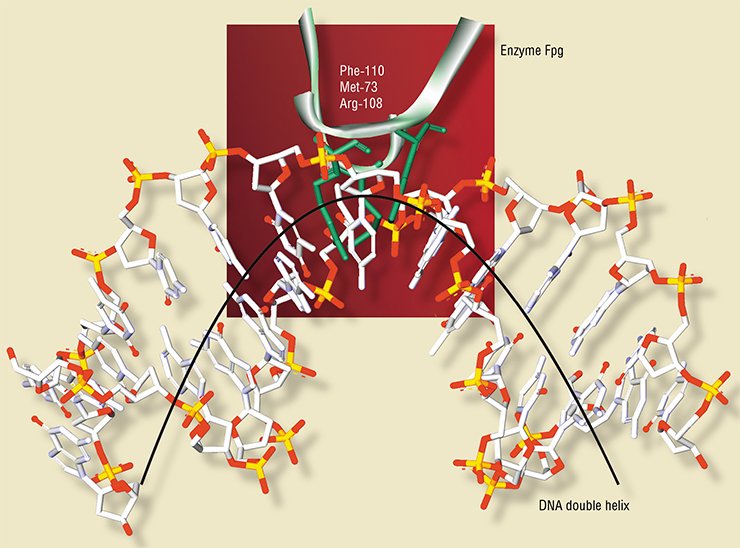
The molecule of Fpg enzyme contains five tryptophan residues. The X-ray structural data for this enzyme in a free state and combined with DNA (Fromme and Verdine, 2002; Gilboa et al., 2002) helped to interpret the recorded dynamics of tryptophan fluorescence during the enzymatic reaction.
This allowed the Novosibirsk researchers to demonstrate that the enzyme interaction with substrates induces conformational changes in the molecules of both the enzyme (for example, insertion of some of its amino acid residues into the DNA double helix) and the substrate.
The change in enzyme conformation (the spatial structure) depends on the type of its interaction with the substrate, that is a nonspecific or specific binding, etc. At least five such conformational rearrangements were observed, each was correlating with certain “movements” in the enzyme molecular structure.
Looking from the opposite side
All the above refers to the enzyme. And what happens with the substrate?
In order to record conformational changes in the substrate, a fluorescent “label”, 2-aminopurine, was inserted into its structure. Actually, the same situation as in the previous case was “filmed” but with another “title character.”
To make the picture more detailed, the DNA substrate was supplemented with two additional “labels” – fluorophore and the so-called quencher (fluorescence extinguisher), which made it possible to record the bending of the substrate molecule by the method of fluorescence resonance energy transfer (FRET).
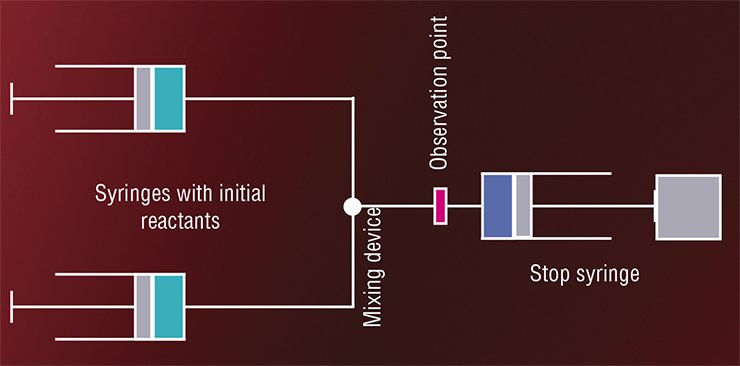
KEEPING THE MOMENT ALIVEAs early as the 1920s, the first so-called flow method was designed: a chemical reaction is triggered by an instant mixing of reactants under flow conditions (Hartridge and Roughton, 1923). In the continuous flow method, solutions of reactants under pressure are put into the mixing chamber wherefrom the mixed solution is conveyed at a constant rate to the tube along which the target concentrations are measured. The “age” of the mixture, i.e. the time from the beginning of the enzymatic reaction is estimated according to the distance from the mixing chamber to the observation point. Unfortunately, this process requires several liters of initial substances, while enzymes are expensive and not easily obtainable products.
In the 1930s—1950s, two modifications of this method were proposed (Roughton and Millikan, 1936; Chance, 1940, 1948). In the accelerated flow version, the solutions in question are put into syringes; the plungers are sharply pushed; and the observation is confined to a fixed point in the vicinity of the mixing device. In this case, the flow rate changes with acceleration, which makes it possible to study solutions of different “ages” at the same point. This method requires rather small volumes of reactants (to 0.1 l).
In the stopped-flow version, the solution, after mixing, is fired into a tube that ends with a plunger. The solution exerts pressure on the plunger and moves it until the plunger is stalled by a stop plate. This stops the flow as well. The concentrations of interest are recorded at a certain fixed point, the time sweep of the recorded parameter giving the kinetic curve of the enzymatic reaction in the range from milliseconds to several minutes.
This method, first proposed in 1934 for the reactions taking about 10 s, was then improved to the time range of several milliseconds and then to even shorter times (Gibson, 1952).
Currently, foreign companies manufacture several devices for measuring fast chemical processes, including those based on the stopped-flow approach, which require minute quantities of the tested substances. Correspondingly, this method is widely used in biochemical research.
At the Novosibirsk Scientific Center, the set ups for studying chemical reactions involving radicals by flow methods were designed at the Institute of Chemical Kinetics and Combustion as early as in the 1960s—1970s. However, these devices required large volumes of reactants and were inappropriate for studying enzymes and nucleic acids.
Today, the Institute of Chemical Biology and Fundamental Medicine, Siberian Branch, Russian Academy of Sciences (Novosibirsk, Russia) is involved in an integrated research into the operation of protective and repair systems of living organisms with the help of flow methods. For this purpose, they use stopped-flow spectrometers manufactured by Applied Photophysics (United Kingdom)
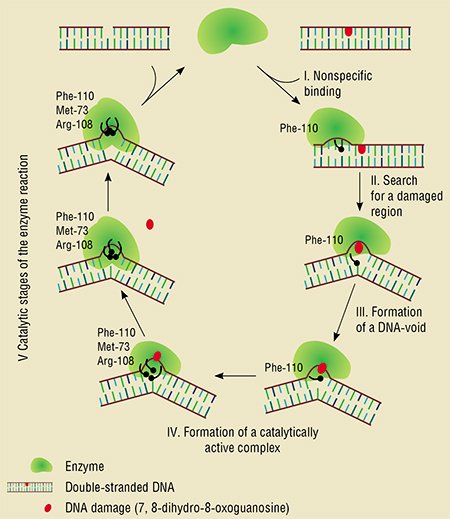 Thus, the researchers obtained two “videos” describing the behavior of the fluorescent elements in the enzyme and in the DNA substrate, which appeared sufficient enough to describe in detail the interaction between the enzyme and DNA. This resulting in detecting mutual conformational transitions during the enzymatic interaction as well as determining the sequence and rates of individual stages in this process.
Thus, the researchers obtained two “videos” describing the behavior of the fluorescent elements in the enzyme and in the DNA substrate, which appeared sufficient enough to describe in detail the interaction between the enzyme and DNA. This resulting in detecting mutual conformational transitions during the enzymatic interaction as well as determining the sequence and rates of individual stages in this process.
These conclusions have been confirmed by other analytical methods such as fast interruption of reaction, which makes it possible to precisely determine the time moments and rates of reaction product generation, as well as mass spectrometry, which validated the formation of the key intermediate products of the enzymatic process.
Thus, state-of-the-art physiochemical methods have made it possible to characterize the nature of conformational transitions that take place during the formation of enzyme–substrate complexes which eventually lead to a catalytically active state of a key DNA repair enzyme.
Such data are of paramount importance for understanding how the retention of hereditary information in living organisms is organized and how genomic integrity is maintained. In general, this study is a good example of how the methods of fast reaction kinetics provide a deeper insight into the mechanisms of enzymatic reactions as well as into highly selective recognition and selective transformation of some kinds of molecules into others.
References
Caldin E.F. Fast Reactions in Solutions. Moscow Mir, 1966. Oxford: Blackwell Scientific Publications, 1964.
Chance B. The Enzyme–Substrate Compounds of Catalase and Peroxides // Nature. 1948. V. 161(4102). P. 914—917.
Koval V. V., Kuznetsov N. A., Ishchenko A. A., et al. Real-time Studies of Conformational Dynamics of the Repair Enzyme E. coli Formamidopyrimidine-DNA Glycosylase and Its DNA Complexes during Catalytic Cycle // Mutat. Res. 2010. V. 685. P. 3—10.
The work was supported by grant no. MK 1304.2010.4 of the President of the Russian Federation, Russian Foundation for Basic Research grant no. 10-04-00070, state contract no. 02.740.11.0079, and by integration projects of the Siberian Branch of the Russian Academy of Sciences nos. 28 and 48