RNA Interference: Fighting Fire with Fire...
“The more” does not necessarily imply “the better”—consider an example of viral proteins. It is the absolute necessity for the infected cell to shut down viral proteins synthesis. The cell follows the homeopathic principle — “like cures like”...
The 2006 Nobel Prize in physiology and medicine was awarded to American scholars Andrew Fire and Craig Mellow for the discovery of RNA interference. This happened to be a surprisingly fast recognition, which followed only 8 years after their publication in Nature demonstrating the possibility of mRNA destruction by means of another RNA molecule, the double-stranded one. An injection of such dsRNA complementary to mRNA coding for the muscle protein unc22 blocked the protein synthesis by the mechanism involving template destruction.
The phenomenon of interference (derived from the Latin, inter—“between” and ferens (ferentis)—“carrying”) is familiar to many of us from the school physics textbook—remember the bright colors of oil patches on the water, the sophisticated pattern of ripples on the water. Besides, this term has general meaningof interaction or superposition of objects similar by nature according to particular rules, and it is widely used in psychology, phonetics, and biologyThe mechanism of mRNA destruction is highly selective: upon injection into the cell, long dsRNAs bind to the cellular enzyme called Dicer, which is a key enzyme in the RNA interference mechanism, whereby they are diced into shorter fragments, the so-called short interfering RNAs (siRNAs). These siRNAs become associated with the cellular complex RISC able to destroy RNA molecules, and target it specifically to those mRNA molecules that are recognized by their complementarity to the siRNA sequence.
As long dsRNAs are not known to be normally synthesized in the mammalian cell, what can be the source of these molecules triggering the RNAi mechanism? One of the possible answers would be the viral infection: many viral genomes are made of such a dsRNA (not of the DNA, as in the majority of living species). The dsRNA can also be an intermediate product of viral replication for a number of viruses. Therefore, the detection of the dsRNA is a red alert signal for the cell, in response to which the cell triggers the defense mechanism destroying the alien genetic information and blocking the synthesis of viral proteins.
The destruction of the viral RNA is not the only defense reaction of the cell as the dsRNAs are able to stimulate the synthesis of interferons and cytokines, thus rendering additional antiviral mechanisms both to the infected and the neighboring cells by activating cell immunity mechanisms and preparing the uninfected cells to virus invasion.
Double-stranded RNAs are involved in the functioning of two important cellular mechanisms—antiviral defense and gene expression regulationRecently, the dsRNA structurally different from the viral RNAs were shown to be synthesized in the cell. These so-called microRNAs are able to shut down the translation of cellular mRNAs involving the RNAi mechanism. Thus, the cell can use the microRNAs encoded in its own genome for the regulation of expression of cellular genes in the course of its developmental program.
The understanding of a variety of cell functions mediated via the RNAi mechanism—from the defense from foreign genetic information to the regulation of cell development — raised the question whether it is possible to use the short synthetic RNAs complementary to the cellular mRNAs for the destruction of the latter. This would yield an invaluable experimental tool for the control of expression of any cellular gene of interest. Moreover, such short oligoribonucleotides are not recognized by the cell as potentially dangerous components of infectious viral genome. They do not activate cellular antiviral mechanisms while targeting solely at the destruction of cellular mRNAs complementary to the interfering RNA sequence.
Experiments proved that this is possible, and currently such synthetic molecules involving the RNAi mechanism are widely used for the regulation of gene expression. They work in a very selective way by destroying only the targeted mRNA molecules and acting at incredibly low concentrations. The development of such a powerful tool opened up new horizons for the researchers in the development of the entire range of drugs suppressing the activity of practically any gene of interest, including viral and tumor genes.
On the basis of small interfering RNAs, siRNA-based inhibitors of important therapeutic target genes, including the genes involved in tumorogenesis, are being developedOnly in the field of oncology, potential targets for interfering RNAs represent a range of mRNAs coding for different classes of protein molecules involved in malignant transformation: cell cycle regulators, angiogenesis factors, proteins involved in the processes of metastasis, ageing, and apoptosis, immunosupressors that hinder the efforts of an organism to fight malignant cells, as well as the proteins responsible for the resistance of cancer cells to chemo- and radiotherapies.
Universal technology
The problem of cancer cell resistance to antitumor drugs is the central issue in the treatment of a number of oncological diseases. Resistant cancer cells can survive and reproduce in the presence of high concentrations of cytostatic drugs usually fatal for “normal” cancer cells.
The syndrome of multidrug resistance is associated with hyperexpression of the cellular gene MDR1 encoding for P-glycoprotein, a membrane transport protein pumping the anticancer drugs out of the cell and reducing their effective concentrations in cytoplasm.
The activity of the molecular pump can be blocked by special chemicals, the inhibitors. However, this does not seem to be the optimal approach to the problem, due to the toxicity of inhibitors, which aggravates the adverse side effects of chemotherapy. The development of drugs eliminating the source of resistance syndrome, i.e., suppressing MDR1 expression, would be an alternative in this situation.
Such siRNA-based drugs were developed at the Institute of Chemical Biology and Fundamental Medicine of the Siberian Branch of the Russian Academy of Sciences in Novosibirsk. The candidate drugs represent the complexes of synthetic oligorybonucleotides sharing homology with different regions of MDR1 mRNA.
In order to select the most efficient siRNA molecules, the cancer cells resistant to cytostatic vinblastin were treated with candidate siRNAs, and only those molecules restoring sensitivity of cancer cells to the cytostatic drug, which leads to the cancer cell death, were selected. Treatment of cancer cells with the most potent of synthesized siRNAs leads to a 20-fold reduction in the amount of P-glycoprotein in target cells within 3 days.
The same approach was employed by researchers of the same institution in order to obtain inhibitors of other cancer-related genes, particularly, myc family protooncogenes, important regulators of the cell cycle. Hyperexpression of c- and N-mycs is known to be one of the causes of neuroblastoma, a malignant tumor made of incompletely differentiated neural cells, which gained an uncontrolled cell division phenotype due to expression control disorder. Those malignancies frequently occurring in children are far from being 100% responsive to treatment.
In a series of experiments involving 21-mer interfering RNAs designed for the suppression of c-myc protooncogen expression, we were able to obtain a highly efficient suppressor molecule, which causes a 20-fold reduction in the amount of mRNA of the oncogen upon introduction into the cell as compared to control. As a result, the relative amount of cancer cell dropped fourfold after 2 days of treatment.
Moreover, one of the obtained siRNAs was able to suppress the expression of both oncogenes, c-myc and N-myc. This molecule turned out to be efficient for treatment of different types of neuroblastomas, including those resistant to standard antitumor medications.
The designed siRNAs can be considered as prototype drugs capable of a multifold improvement of the efficiency of antitumor chemotherapies. Obviously, before being introduced into clinical practice, these drugs still have a long way of in vivo experimentation using lab animals, preclinical, and clinical trials.
The beneficial chemistry
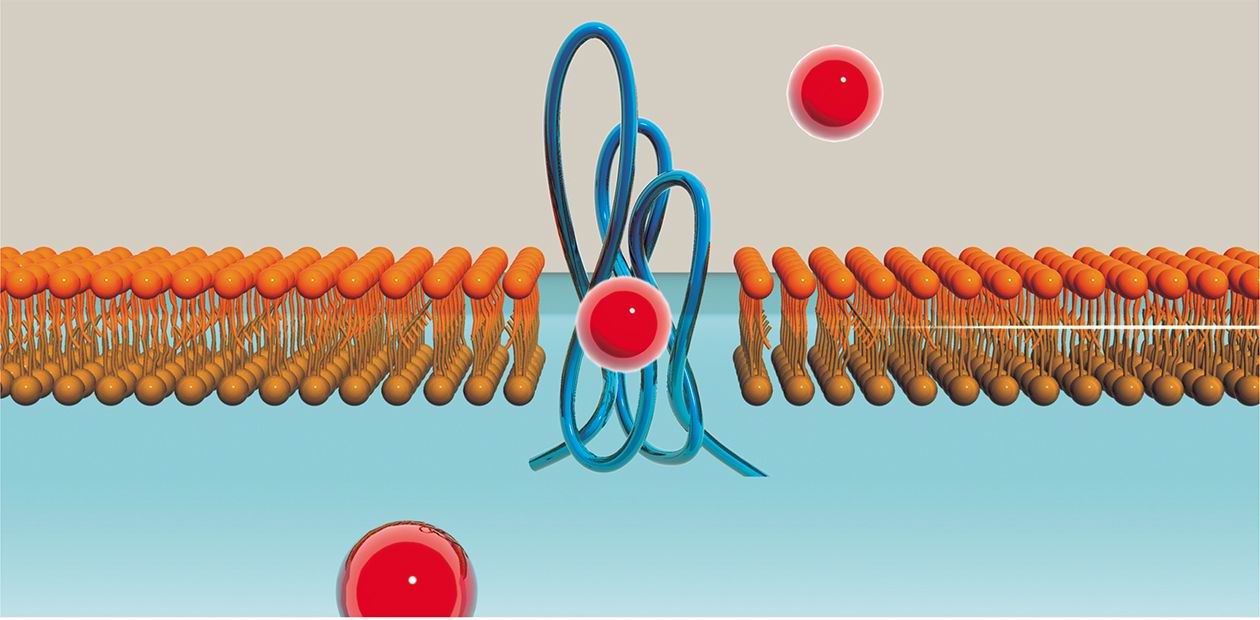
In order for siRNA to become a real medication, two problems are to be resolved: siRNA protection and delivery.
The first problem results from intrinsic instability of RNA molecules in blood and other body fluids, which seems to be quite a natural property assuming the role of RNA as a genetic information carrier, the amount of which should be easily controlled. Intra- and extracellular RNAses actively degrade RNA molecules, working as such a regulator. In order to protect siRNA from RNAses, the naturally occurring nucleotides comprising these molecules are being replaced for their chemically modified analogs rendering interfering RNAs RNAse-resistant. However, this approach might happen to be a double-edged weapon as the biological activity of siRNA might be compromised or completely lost. Search for the nucleotide analogs is therefore a global research effort. The algorithm for the design of nuclease-resistant interfering RNAs developed at the Institute of Chemical Biology and Fundamental Medicine is based on nuclease-sensitive site mapping and directional protection of those by chemically modified nucleotide analog substitution. Such addressed protection results in an 8-hour biological activity lifetime in the presence of 5% serum for a protected siRNA versus 5 minutes of non-modified siRNA lifetime whereupon it is completely degraded. Due to this acquired stability, the frequency of injection of a new dose required for the maintenance of biological effect reduces dramatically.
Search for novel “carrier and packaging” molecules for siRNA delivery is carried out in labs all over the world, and Russia is no exception. In the nearest future one can expect breakthrough results which would possibly start a new era in drug industry technologiesThe other problem is the design of delivery vehicles able to address the drug to the targeted cells and tissues. Addressed delivery of siRNA-containing complexes is achieved by the introduction of specific molecules for recognition of different cell types. As an example, antibodies with their highly specific binding represent one of the targeting molecules of choice.
The cell membrane presents another barrier, which is not easily crossed by any charged molecule, including oligoribonucleotides. To overcome this obstacle, the use of cationic lipids and polymers as delivery vehicles might be of help. They aggregate with siRNA molecules into specific-size complexes able to cross the cell membrane via cellular transport machinery. Yet another promising approach is based on linking siRNA to natural molecules, which has been shown to be naturally capable of traversing the cell membrane, such as cholesterol, folate, and other molecules necessary for cell metabolism.
Delivery of interfering RNAs to the particular cell types and tissues has been already successfully resolved, and the efficient shutdown of target gene expression has been demonstrated in lab animal models. However, experts consider the current level of developments in this field as a promising but modest achievement. It is the design of appropriate vehicles and protocols for the delivery of RNA-based drugs into a wide range of target cell types (and for different disorders) that the fast introduction of RNA-based therapies into clinical practices and the demonstration of its power and possibilities will depend on.
References
E.B.Logashenko, A.V.Vladimirova, A.N.Zenkov, M.N.Repkova, A.G.Veniaminova, E.L.Chernolovskaya, and V.V.Vlassov (2005) Reversal of multidrug resistance phenotype by small interfering RNAs // Bulletin of the Russian Academy of Sciences, Chemistry, 2, 41—44
Aronin N. (2006) Target selectivity in mRNA silencing. Gene Therapy, 13, 509—516.
Corey D. (2007) Chemical modification: the key to clinical application of RNA interference? // The Journal of Clinical Investigation, 117, 3615—3622.
Grimm D. and Kay M. (2007) Therapeutic application of RNAi: is mRNA targeting finally ready for prime time? The Journal of Clinical Investigation, 117, 3633—3641.
Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S. E., and Mello C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans // Nature, 391, 806—811.