RNAI and Antisense Technology: Rivals or Partners?
On their first encounter with RNA interference technology, then a newcomer in the toolbox of efficient gene knockout methods, many research and medical professionals believed it to become a complete alternative to the traditional antisense approach. The vivid discussion stimulated a wide coverage by Nature in 2004: a series of publications debated over possible rivalry between RNAi and antisense
On their first encounter with RNA interference technology, then a newcomer in the toolbox of efficient gene knockout methods, many research and medical professionals believed it to become a complete alternative to the traditional antisense approach. The vivid discussion stimulated a wide coverage by Nature in 2004: a series of publications debated over possible rivalry between RNAi and antisense.
Ardent supporters of the new approach argued that RNA interference had already achieved the heights that the antisense approach had never even dared to approach. They referred to their research results confirming the new method to be orders of magnitude more sensitive and efficient than the older one. In accordance with some estimates, an efficient silencing concentration for siRNA is 100—1000 times lower than for antisense oligonucleotides.
The followers of traditional antisense approach insisted that the efficiency of both methods is comparable at least for expression shutdown in cell cultures. In their opinion, the underestimates of antisense oligonucleotide efficiency result from the use of first generation oligonucleotides for comparative studies (while more advanced third generation oligonucleotides were at hand) otherwise incorrect selection of target sequences might have been the cause.
As far as in vivo application of RNAi was concerned, a representative of ISIS Pharmaceuticals biotech company argued that a considerable effort for optimization of this approach was required in order that it could at least match the antisense drugs. This estimate was not unexpected, as ISIS was the only company to succeed in entering the pharmaceuticals market with their antisense-based drug Vitravene used for treatment of skin diseases.
However, this opinion would hardly be shared by another representative of the pharmaceutical industry. From the viewpoint of vice-president of SIRNA Therapeutics, the RNAi breakthrough came out of the blue, yet, for his business that was an opportune surprise. His company, known before 2003 as Ribozyme Pharmaceuticals, was in a deep crisis since its most promising antisense-based drugs had proven inefficient in clinical trials. Upon refocusing on RNAi technology, it re-entered the market as SIRNA Therapeutics. Their Sirna-27 drug aimed at age-related macular degeneration treatment has become the first RNAi-based preparation to start in clinical trials.
Interestingly, other pharmaceutical companies which employed alternative approaches to drug design including the antisense technology, have never considered the siRNA technology as a rival. Taking advantage of their rich RNA chemistry experience, they immediately entered the siRNA-based drug race. Their experience in the design of delivery vehicles to different cell types turned out especially valuable as the lack of these hampers the transformation of both technologies into the full-scale drug design and manufacture industries.
Companies specialized in delivery vehicles usually optimize their products for both types of drugs, as it is done by the Illinois company Neopharm for their lipid delivery systems. Their head of research believes that, by and large, siRNA drugs work better than the antisense-based ones.
Besides, the delivery problem is not the only field where the siRNA industry requires “external” experience. There are some problems where only a combination of RNAi and the antisense technology can yield a solution. For example, siRNAs are efficient tools for gene silencing. Is there a way to make gene expression more active?
Age-related macular degeneration or vision disaster
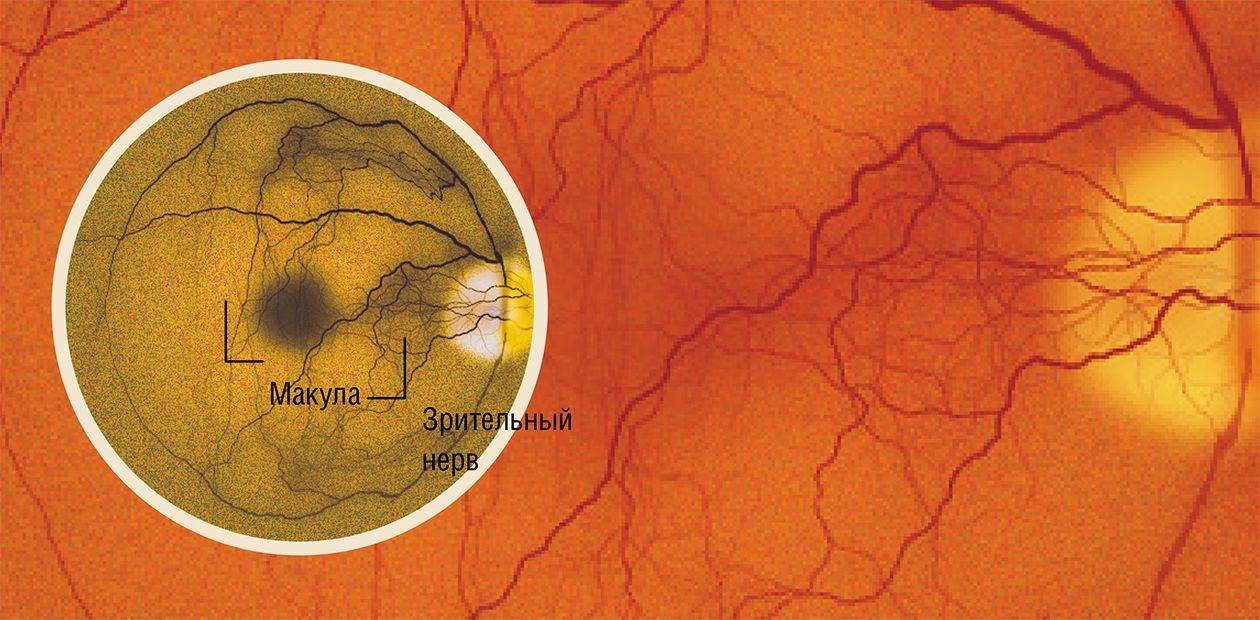
Macula (macula lutea or the yellow spot) is the central region of the retina (about 5 mm in diameter) consisting of the most sensitive cells. Healthy macula is responsible for central vision as well as for about 80% of the overall visual acuity, the remaining 20% being accrued due to the activity of the rest of the retina.Macular degeneration is the major cause of irreversible vision loss in aged patients. A common diagnosis for 70—80-year old patients in the past, currently macular degeneration is breaking this age limit down to 40 to 60-year old. According to the American Association of Ophtalmology survey, the age-related macular degeneration (AMD) accounts for the majority of vision loss cases in patients above 50 in the United States.
The early symptoms of developing AMD appear as punctated defects in the field of vision, with wider areas of the retina subsequently involved leading finally to a complete loss of central vision. One of the early symptoms of the disease is vision distortion (metamorphosia), which can appear as a wavy pattern of a square grid placed with some fields of vision completely disappearing. An easy and efficient tool for metamorphosia detection is Amsler Grid Test consisting of a central black dot with a superimposed grid.
Age-related macular degeneration can lead to a complete blindness. The most threatening symptom in patients with partial vision loss is their inability to see faces – either completely or at close range. SIRNA-27 is the first of siRNA-based drugs to enter clinical trials. Out of a total number of 8 siRNA drugs in trials, three are aimed at AMD treatment
It is now known that a number of diseases including the oncological ones are caused by overexpression of some cellular microRNAs, which in turn results in down-regulation of particular genes. MicroRNAs are the cellular analogs of SiRNAs produced by the cell itself for the regulation of its gene expression. The so-called microRNA antagonists (antagomiRNAs), the antisense molecules designed to inactivate cellular microRNAs, can be used in order to lift that microRNA blockage of gene expression.
There are some other applications where antisense oligonucleotides may prove to be more efficient than interfering RNAs. For example, some viruses can present a die hard target for the latter. The original viewpoint considering the RNAi mechanism as an intrinsic cellular antiviral immunity turned true for plant and lower animals only. Although since the beginning of RNAi technology applications data have been communicated on successful suppression of HIV-1, HBV, HCV, SARS, and influenza type A viruses in different experimental systems, many viruses have appeared to employ active and passive anti-RNAi strategies. Many virus-encoded proteins are RNAi suppressors, including NS1 protein of influenza A virus, HCV envelope protein, HIV-1 Tat protein, VP35 protein of Ebola virus, and others.
Altogether, many RNA viruses turned out to be well protected against RNAi. The retroviral genome (for example, Rous sarcoma virus) is safely packed inside the viral core being merely inaccessible for RNAi. HIV-1 belongs to the genus of lentiviruses, Retroviridae family, and its principle of virus assembly is very much similar to classical retroviruses.
The data concerning the inactivation of HIV-1 genome inside such a protein shell by siRNAs are controversial. In case the HIV-1 virus attacking the cell proves safeguarded from RNAi, the idea of anti-HIV preventive vaccine development will have to be discarded. Upon HIV-1 virus insertion into the genome of attacked and initiation of viral genes expression, interfering RNAs will only have to struggle with multiplying copies of viral RNA genome, thus trying their best to harness viral replication, but complete clearance of the virus out of the cell will be impossible.
Besides, almost all of the viruses exploit an escape variant strategy for passive avoidance from RNAi control. Viruses mutate at incredible rate, thus accumulating mutations in their genome. Substitution of a single nucleotide in a portion of the viral genome complementary to interfering the RNA sequence may be sufficient to “inactivate” siRNA. Antisense drugs are much less demanding in that respect, and they can tolerate several substitutions without complete loss of antiviral activity. While for interfering RNA drugs the occurrence of escape variants has been demonstrated for most dangerous health-related viruses, only SARS have been shown to be able to avoid (though incompletely) antisense drug suppression.
Currently, it seems clear that RNAi is not a rival aiming at driving the traditional antisense technology out of the pharmaceutical market. Both approaches have their supporters and critics, and there are problems more successfully tackled by one or the other technology. Also, there are problems more easily resolvable by combinations of antisense and interfering RNA drugs.
There is another question, more difficult to answer: will the originally super-successful RNAi technology share the destiny of the classical antisense approach, which did not meet the high expectations it inspired two decades ago?
The lists of “mass-murderers”, the most dangerous enemies of contemporary medicine, which can theoretically be treated by antisense-based or siRNA-based drugs, are very much similar. They include all kinds of oncological diseases and neurodegenerative disorders, such as Alzheimer`s and Huntington`s diseases, the most important cardiovascular (hereditary coronary disorders, hypertension, atherosclerosis, myocarditis, heart hypertrophy, and infarction) and immune (asthma, psoriasis, allergic disorders, inflammation) diseases, as well as a number of virus and virus-unrelated hepatitis, and other viral infections.
The roadblocks on the way to clinical application of both technologies also seem alike: potential drug toxicity, off-target reactions (unexpected shutdown of genetic pathways that were not supposed to be silenced), and the above-mentioned problem of therapeutic RNA delivery to different types of cells and tissues.
As an experimental gene knockout technology, RNAi has also proved to be a successful approach of first choice for a widest range of applications. It has another advantage of reducing the costs of candidate drugs testing in pharmacology. Will siRNA-based drugs form the foundation for a new generation of therapeutic drugs or repeat the fate of antisenses?
Most specialists are pointing out, though guardedly, at a possibility of ex vivo therapeutic application of siRNAs, i.e., their use for modification of cells isolated from the patient. At the same time, the estimates of industry representatives tend to sound more optimistic than those from their academic colleagues believing that RNAi will never get out of the Petri dish before the delivery problem has been resolved.
References to the Editorial comment
Dykxhoorn D. M. and Lieberman J. The silent revolution: RNA Interference as Basic Biology, Research Tool, and Therapeutic // Annual Review of Medicine, 2005. V. 56(1):401
Haasnoot J., Westerhout E. M., Berkhout B. RNA interference against viruses: strike and counterstrike // Nature Biotechnology 25, 1435—1443 (2007)
Sassen S., Miska E. A., Caldas C. MicroRNA—implications for cancer// Virchows Arch. 2008 January; 452(1): 1—10
Pellish R. S., Nasir A., Ramratnam B. and Moss S. F. RNA interference—potential therapeutic applications for the gastroenterologist // Alimentary Pharmacology and Therapeutics 27 (9) , 715—723, May 2008
Wu W., Sun M., Zou G.-M. and Jianjun Chen. // International Journal of Cancer: 120, 953—960, 2006
Marquez R. T. and MCcaffrey A. P. Advances in MicroRNAs: Implications for Gene Therapists// Human Gene Therapy.January 1, 2008, 19(1): 27—38. doi:10.1089/hum. 2007.147.
Wu L., Belasco J. G.. Let Me Count the Ways: Mechanisms of Gene Regulation by miRNAs and siRNAs // Molecular Cell 29, January 18, 2008:1—7
Kurreck J. Antisense technologies. Improvement through novel chemical modifications // FEBS Journal, Volume 270, Number 8,April 2003 , pp. 1628—1644(17)
Clayton J. The silent treatment. // Nature. Vol 431, 599—605, 2004
Lieberman J., Song E., Lee S. K., Shankar P. Interfering with disease: opportunities and roadblocks to harnessing RNA interference // Trends in Molecular Medicine 2003 Sep; 9(9):397—403
Tang Y., Ge Y.-z., Q Yin J. Exploring in vitro roles of siRNA in cardiovascular disease // Acta Pharmacologica Sinica, Volume 28, Number 1, January 2007, pp. 1—9(9)
De Fougerolles A. R. Delivery Vehicles for Small Interfering RNA In Vivo // Human Gene Therapy 19:125—132, February 2008
Li C. X., Parker A., Menocal E., Xiang S., Borodyansky L. and Fruehauf J. H. Delivery of RNA interference. Cell Cycle 5(18):2103—2109, 2006 September 15
Love T. M., Moffett H. F., Novina C. D.. Not miR-ly small RNAs: big potential for microRNAs in therapy // Journal of Allergy and Clinical Immunology, 2008 Feb ;121 (2):309—19
Aboul-Fadl T. Antisense Oligonucleotides: The State of the Art // Current Medicinal Chemistry, Volume 12, Number 19, September 2005 , pp. 2193—2214(22)