We and our microbes
The very phrase “We and our microbes” testifies to the established anthropocentric view on the microscopic inhabitants of our body – the microbiota. However, microbiota is an indispensable part of our organism: it develops and functions, just like other organs and tissues. That is exactly why scientists speak of the “superorganism” – that is, that each of us is a complex ecosystem. Even though the microbiota has been studied for many years, its story is far from closure. The number of things unknown and uncomprehended is far greater than everything we have been able to discover – but even the current results are astonishing
Geoffrey Gordon, an American professor and scientist who has been called ‘the father of microbiome’, was nominated for the Nobel Prize in 2015 for his research on the interaction of intestinal bacteria with the intestinal wall and the human organism as a whole, which is another proof of the importance of scientific results in this area. What is the microbiome (or microbiota)? Why is it difficult to establish who interact with who, and how?
An amazing fact: the number of microorganisms inhabiting our body (viruses, bacteria, and fungi), with most of them inhabiting the gastrointestinal tract, exceeds the number of our own cells. The most striking thing is the number of genes – on average, there are 500 times more microbial genes in our organism than human genes.
The term ‘microbiota’ denotes the sum of all microorganisms (bacteria, fungi, and viruses) that inhabit the human body. The resident, or obligate microbiota consists of organisms always present in the specific part of the body. This community, which has formed during the mutual evolution of microbes and their macroorganism host, is symbiotically connected to the latter and often play protective and other useful roles. For the microbiota, this is a way to procure a stable environment. Along with the resident microbiota, there is also the transitory microbiota – “foreign” microorganisms are incapable of long-term survival in our body.The total number of microbial cells in the human body is believed to reach 1014, which is an order of magnitude greater than the number of our own cells. The bulk of the microbial population inhabits the large intestine (over 500 species), followed by the skin (104–106 per cm2), conjunctive, upper respiratory tract, and vagina
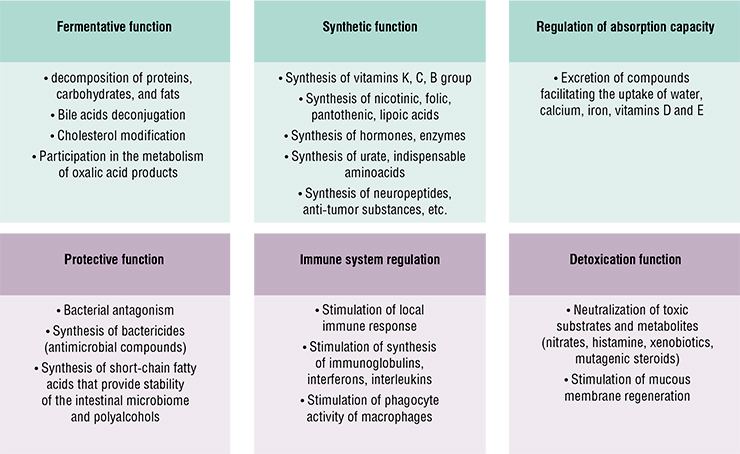
Microorganisms perform functions essential for us, including trophic and regulatory functions. For instance, intestinal bacteria decompose undigested food fragments and excrete various compounds into the intestinal lumen, including vitamins, enzymes, and other biologically active compounds. Microorganisms actively participate in metabolic processes, including detoxication, by decomposing toxins that come in with water, food, air, etc.
The total weight of our microbiota is around 2 kg, which is comparable to such major organs as the brain or liver. Unlike our organs, it is very poorly studied, especially given its extreme variability from person to person and depending on a person’s condition. The key mystery is how it switches from its normal state to a pathological one.
Gaining weight out of air
In the modern post-industrial era, disruptions of the established microbial balance of the human body are a common thing. This not only leads to discomfort but also to untimely aging and development of various pathological conditions, which affect way more than just the digestive tract. As for the intestine, in the past fifty years, the frequency of its diseases, including oncological diseases, has exploded, with colon cancer leading statistics in developed countries. This is topped by inflammatory diseases, such as various types of colitis and Crohn’s disease.
There is an obvious microbial imbalance here that affects not just individuals, but whole populations. It is a paradox: modern people eat better, and our diet is more diverse than ever before, yet almost every person has some signs of digestive dysfunction.
It turns out that the key is not in the amount of food, or even calories, that a person consumes. Depending on the enzymatic properties of specific classes, genera and species of bacteria, our microbiota is capable to use even those compounds of food that were previously considered indigestible. There is a saying that some people get fat from air. This means that their bacteria extract extra calories from such compounds and share them with the host’s body, which launches several processes that lead to obesity.
From a chemist’s viewpoint, the digestive tract is a unique structure. The stomach and small intestine, locked by sphincters, are like large chemical reactors where the process of digestion occurs, powered by an assortment of chemicals such as enzymes and strong detergents, including cholic acids excreted with bile. The acidic environment of the stomach, with the lack of oxygen in the small intestine, create relatively sterile conditions. At this stage, easily digestible metabolites are extracted from food: fats, amino acids, or short peptides as well as “fast” carbohydrates.
The upper sections of the gastrointestinal tract have meager bacterial flora, and this is not by chance. The acidity combined with antibacterial enzymes of the mouth cavity and some immune factors control the proliferation of microorganisms just enough to control the rotting processes in the food. If the number of microbes in the small intestine increases, it is accompanied by pathological reactions. Not only do bacteria consume the “easy” metabolites, but they also excrete toxins, which are instantly absorbed into the bloodstream.
Whatever is left, goes into the colon. By the time it reaches the lower sections of the small intestine, where it is connected to the colon, almost all bile, which is toxic to microorganisms, has been completely resorbed. That is why the colon, with its relatively neutral environment and higher oxygen concentration, is home to an intricate and intimate cooperation of various bacteria.
Is there such a thing as normal flora?
Recently, the microbiota of different mucous membranes of humans has been under study as a potential pharmaceutical target. The shifting philosophy of modern medicine, with predictive and preventive function coming into the light, gives hope that this new branch of biomedical research will let us prevent or even cure certain diseases.
To influence our microbiota specifically, we should know what normal microbiota is like. The very concept of normal microbiota is rather illusory. A person is fine if he/she feels well and does not experience any digestive problems.
If we look deeper, normal microbiota is such a condition of the trillions of bacterial cells when, first, there are no inflammatory reactions in the body, and second, the range of metabolites produced by the microbiota is sufficient for its long-term autonomous existence without any bacterial intervention from the outside.
There are ongoing debates on whether this microbial kingdom can be divided into distinct enterotypes, or rather, it is a spectrum of dynamic states that transition into one another and defy the distinct classification.
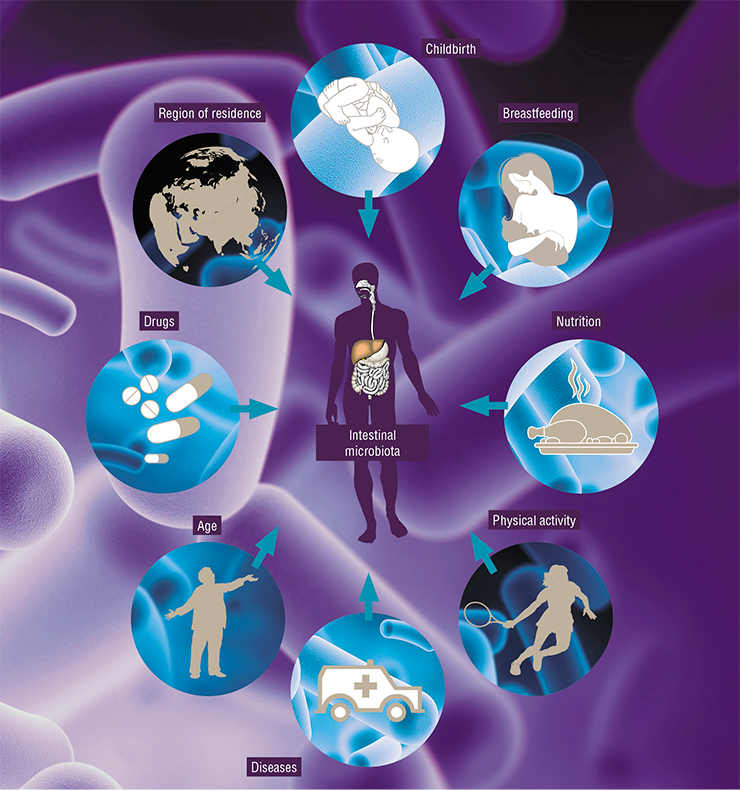
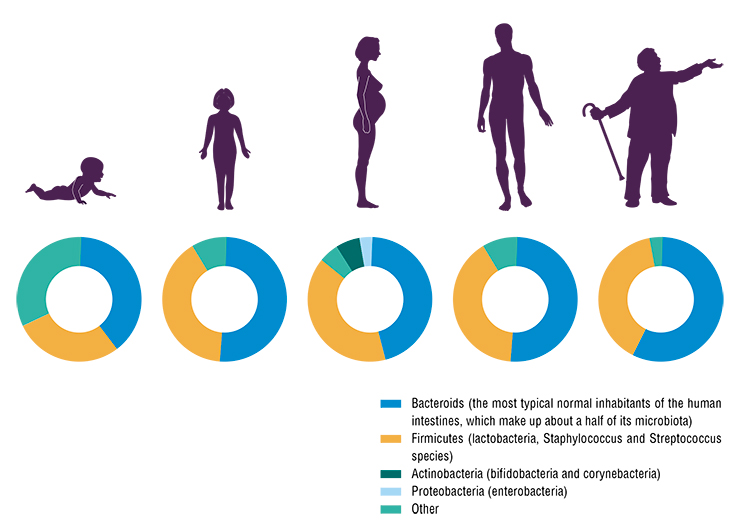
Many factors affect the microbial community. Overall, high diversity of microbes and their genes in the intestines is a good sign. With age, the diversity of an individual’s intestinal microbiota drops. Genetic estimates of microbial communities show that their number in the old and sick is halved compared to the healthy and young. This leads to metabolic syndrome, diabetes and other diseases.
The human microbiota also differ from country to country. This is an issue of context. If you compare major cities in Europe and Russia with populations above one million, you will not find any substantial differences. However, in remote areas far from big cities, the microbiota is more diverse and similar to the microbial communities of people who lived 100 to 150 years ago. This means that when we consume less of the so-called industrial foods, our micro-world tends to be more diverse.
The important thing is that we have learned to change the bacterial dynamics. Not only the contents of the microbiome are of interest, but rather its reactions to certain events and activities, such as travel, change of diet, or hospitalization. Like all other cells of our body, the microbiota accumulate mutations. With our own cells, it often results in oncological transformations, but we are still far from understanding what happens to the microbiota. We know that the levels of bacterial mutagenesis correlate with the frequency of use of antibacterial drugs. A glance at the antibacterial policies in different countries – China, the United States, or European countries – shows a clear correlation with the number of mutations accumulated in microbes.
Metagenomic studies are underway – scientists are looking for patterns connecting the presence of specific microorganisms in the microbiome with specific diseases. For instance, Chinese researchers demonstrated a connection between the presence of particular bacteria and the development of type 2 diabetes. At the moment, this is just a formal description of the phenomenon, and its mechanism remains unclear.
One should not forget about the ecological component of the microbiome. In nature, not only the human genes, which are passed down from the ancestors, are synchronized; so do the bacterial genes, which are transferred in human populations both vertically and horizontally. For instance, most mammals are coprophagous – this behavior is inborn. This reflex is present in primates, including humans, however, children are taught not to follow it at early stages of nutritional learning. This is justified from the sanitary viewpoint, but not so much if you consider the importance of bacterial transfer.
“Store” foods contain many genes of antibiotic resistance, most animals that make up our livestock come into contact with antibiotics one way or another. These substances are used to enhance growth, and cut losses from infectious diseases, which is important when animals are kept in crowded conditions. We humans also often use antibiotics for various diseases, and not only bacterial diseases.
As the result, genes of antibiotic resistance are gradually accumulated in the environment. One of the first research articles on this topic demonstrated that the soil contained most modern, yet not chemically synthesized antibacterial compounds, which are one of nature’s ways of inter-bacterial communication.
The most giant reservoir of antibiotic-resistant genes is humankind. Many infectious diseases, such as emphysema, different types of chronic pneumonia and bronchitis, which used to be considered hospital-borne diseases, are in fact the result of bacteria from “natural” habitats within our body entering the bloodstream. By moving, for instance, from the colon to the lungs, such microorganisms can cause illness or even death.
Ideally, to change a combined course of antibiotics, patients should be screened for antibiotic resistance. Otherwise, we will end up with superbugs, like other countries – i. e., bacterial strains insensitive to antibiotics.
Show your metabolites
One of the optimal ways to evaluate the microbiota is by analyzing the range of bacterial metabolites. However, it is not that simple, as the latter are a vast and poorly studied spectrum.
One of the problems is the lack of reliable ways to separate endogenic metabolites, which are produced by human cells, from those that enter the body with food, water and air, and those created by microorganisms. In animal models, this can be done using radioactive or isotope markers; however, it is unacceptable for humans.
Nevertheless, there are groups of metabolites that can be recommended for medical use. First, these are short-chain fatty acids. Their detection is routine: any patient with digestive problems can have the proportion of these compounds tested even before getting a diagnosis.
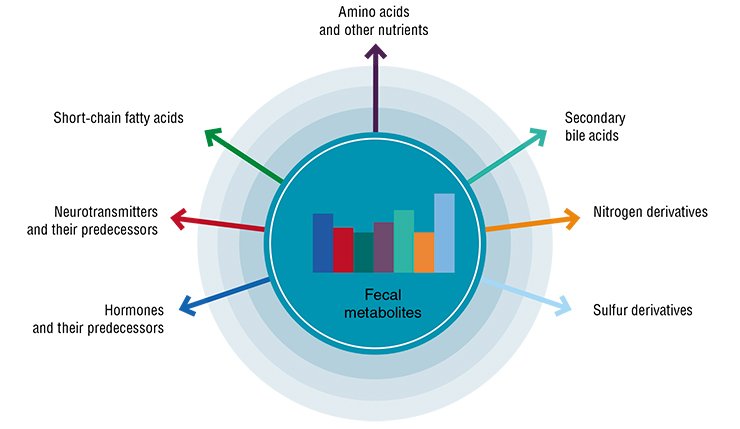
The second group is hormone-like compounds derived from folic acids. Depending on the spectrum of bacteria that oxidize either the products of adducts of the folic acid with glucuronides or the folic acid itself, different active compounds from the steroid series can form, which have specific receptors on the adrenal glands, pericardium, myocardium, lungs, etc. However, nowadays endocrinologists simply cannot “see” these substances.
The third group is indoles, which, depending on their chemical modification, can either have a protective effect by suppressing dangerous intestinal pathogens, or be a powerful modulator of anti-inflammatory responses.
What are the feedback mechanisms between the body and its microbiota? When you go on a diet, your E. coli population secretes peptides similar to protein-releasing factors, which are produced in the central nervous system and are used in appetite regulation. When these hormone-like bacterial substances enter the bloodstream, we feel hunger. This is how bacteria “force” you to feed them.The second example deals with people who normally consume few animal proteins. They are likely to have a deficit of digestive enzymes – proteases and lipases in particular – in their small intestine. If such a person forgets to go easy on grilled steak on a picnic, the protein bodies will enter the large intestine, where they cannot be used due to the lack of proteolytic enzymes. This launches intensive putrefaction processes in the large intestine caused by massive proliferation of anaerobic putrefactive bacteria. The next morning the person will feel unwell – a condition similar to a bad hangover due to auto-intoxication
The balance of these substances is important both for selecting personalized treatment schemes and for understanding of how good the final result is, – a well-stabilized physiological state of the patient.
It looks like scientists have a lot of work before them on creating a medically oriented registry of bacterial metabolites, developing inexpensive methods of their registration and usable means of their interpretation by medical professionals. In other words, it is about working out the “indications”. Measuring metabolites is easier and more accurate than defining the makeup of the patient’s microbiome every time (the latter is more important for basic science than for applied medicine).
Dysbacteriosis – once again
In Russian pediatric practice, the diagnosis “dysbacteriosis” is still widespread, even though there is no such disease in the official international classifications of diseases, such as ICD‑10 (The International Statistical Classification of Diseases and Related Health Problems. 10th revision). If a child has diarrhea or other problems with digestion, their stool sample is taken for culture analysis, and the bacteriological laboratory counts the numbers of 20 to 30 microorganisms (bifido-, lacto-, actinobacteria, etc.). The main goal of such analysis is to define whether there is an inflammatory process in the intestines. However, there are cases when the results clearly indicate dysbacteriosis, and yet the patient feels fine and there are no clinical symptoms whatsoever; sometimes, the situation is opposite.
The word “dysbacteriosis” itself is not exactly accurate. There is an imbalance, but not just of the numbers of bacteria, but rather of a large number of metabolic parameters, including the content of short-chain fatty acids, indoles, and free human DNA in the stool (if there is diarrhea, i. e., an inflammatory syndrome, the intestinal epithelium is actively shed). The imbalance manifests itself not only within the bowel, as previously thought – it is a systemic disorder.
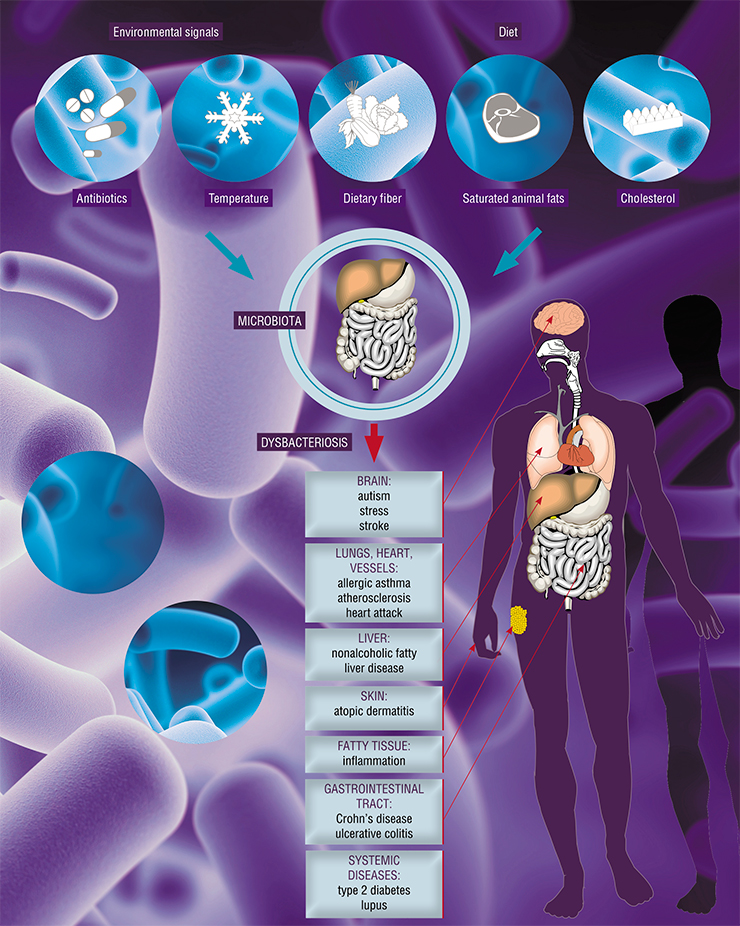
An imbalance of intestinal microflora is connected to the development of oncological diseases, different forms of inflammatory processes, and clostridial infection (Clostridia are present in the bowel in small numbers and usually do not cause any pathologies). As a rule, clostridial infections are psychogenic; these cases tend to be serious and are treated with long-term aggressive courses of antibacterial drugs.
During the first enthusiastic efforts of gene sequencing and locating bacterial associations, it turned out that there was a whole range of different diseases connected with the intestinal microbiota. They include the metabolic syndrome, type 2 diabetes, asthma, and autism. We know that mice who have received transplanted microbiota of a patient with autism develop a similar syndrome, which can be relieved by transplanting microbiota from healthy donors.
Crohn’s disease
Let us take a closer look at Crohn’s disease – a chronic inflammatory disorder of the bowel. In the past century, the epidemiology of this serious disease has taken a turn for the worse: from 2—3 cases per 100 000 people in the beginning of the last century to almost 150 in some Scandinavian countries. These statistics include only cases with distinct clinical symptoms. Methods of capsule endoscopy, which have given us the possibility to examine the middle of the small intestine, will probably make this statistics even worse.
There have been numerous attempts to explain the reasons and mechanisms of Crohn’s disease. There is a genetic theory for cases with purely autoimmune nature. Today, we have the tools for rather accurate recognition of the genetic patterns that correlate with the probability of Crohn’s disease in a specific patient.
Historically, Crohn’s disease has been associated with many infectious diseases, such as intestinal tuberculosis. Recently, French researchers studying the Western European population discovered a specialized E. coli strain, LF82, which is closely associated with Crohn’s disease. This well-known uropathogenic strain causes cystitis (bladder inflammation) and tends to have higher adhesion and invasion to epithelial cells. It turned out that in some patients with Crohn’s disease, this E. coli strain makes up to 80 % of the microbial communities surrounding intestinal ulcers (normally, this concentration is within 3 %).
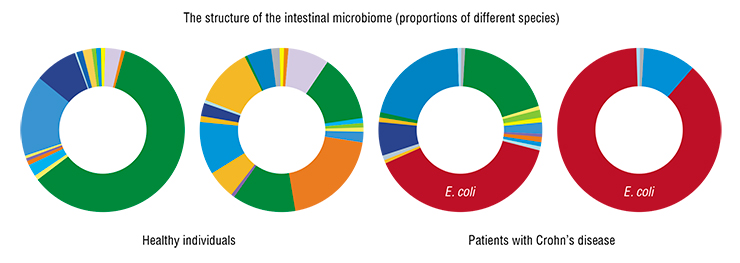
One might assume that with the culprit pathogen known, there would be at least some advances in treatment. However, after performing a phylogenetic analysis, i. e. isolating and characterizing microbial strains from Russian patients, we realized that “our” E. coli is extremely diverse phylogenetically and may be a part of the normal symbiotic human microflora.
What we have found is a sequel of the “philosophy of contexts”. In all E. coli strains being studied from patients with Crohn’s disease, we found operons (functional genomic units of bacteria) connected with the use of propanediols – products of decay of fucose, which is a part of the protective mucus excreted by the intestinal mucous membrane.
These and other peculiarities of E. coli (such as the lack of genes coding iron-containing antioxidant proteins) give it advantages over other microbial strains present in the bowel, which is similar to some models of human societies when under certain conditions, obscure groups of people become the society’s strongest and main beneficiaries.
We are talking about a new hypothesis, according to which in certain scenarios connected with the use of antibiotics, dysbacteriosis, chemical intoxication or chronic inflammatory processes in the mucous membranes, E. coli, while not itself a pathogen causing a specific disease, gets extended possibilities for proliferation, and its numbers become overwhelming. This launches a pathological process.
What is FMT?
Few people know about a procedure called fecal microbiota transplantation (FMT). Recently, FDA decided to suspend the use of FMT in clinical practice, quoting two cases of complications (one fatal) after fecal transplantation. The reason was that patients were infected with E. coli strains with genes of multiple drug resistance to antibiotics.
In Russia, fatal complications of appendicitis make up 1.5 % of all cases, even though it has been a routine surgical procedure for a long time. All published FMT protocols combined yield a complication rate of less than 0.001 %. Moreover, we are speaking about patients after oncological surgery and chemotherapy who were apparently weakened. For instance, our colleagues from the Raisa Gorbacheva Memorial Research Institute of Children Oncology, Hematology, and Transplantology in St. Petersburg are saving children with acute leukemia, where sepsis is the main complication, with the help of fecal transplantation. Using FTM in dire situations without alternatives, when the immune system is virtually nonexistent, and antibiotics do not work, can save up to 60 percent of the patients.
However, it is necessary to work out clear indications for the use of FTM. It is an issue of context: if we perform a procedure, having evaluated all balancing factors in a specific patient, we have a solid chance of helping them.
Manipulations with microbiota are one of the promising directions in personalized medicine. Just like the use of target drugs, which have side effects, it must be a thoroughly thoughtful step. To evaluate all possible relationships between the microbiota and the host organism is next to impossible: they are so many that they have not even been properly cataloged yet. However, developing the so-called surrogate evaluation techniques based on metabolic profiling, endoscopy and certain methods of control is a feasible goal.
Our own observations of the FMT and colonization of the intestine with “foreign” bacteria show that just like with the blood type, there are matching groups of donors and recipients. That is why even a clinically successful procedure may provide only temporary results as the transplanted flora eventually “fades away”. There is ongoing research aimed at the creation of artificial feces, or artificial microbiota – but with limited success.
The idea that we are not alone on our life path has been around for a long time, but it was only in the current century that scientists and medical experts got the necessary tools to study communities of our microbial co-inhabitants.
The very issue of exhaustive cataloging of all microorganisms inhabiting the human body is primarily technological, as few of these organisms can be grown on artificial media. There are several reasons for that. Even the most advanced growth media can lack components indispensable for the life cycle of a particular strain. Moreover, different bacteria rarely live solitarily and usually form communities, thus supporting and supplementing each other.
However, to discover whether there are specific bacteria in a sample, it is enough to extract nucleic acids, especially DNA, and “decode” them to compare with existing databases. This procedure is becoming less and less expensive, which has made it possible to study the great variety of microorganisms that coexist with us.
The most complex and the most interesting study target in this sense is the intestines. The intestinal microbiome is a complex and hierarchically organized structure. When they speak of trillions of intestinal bacteria, they usually mean the microflora of the intestinal lumen. However, there are microcolonies of bacteria nested within the mucous membrane proper – i. e., in the layer of epithelial cells lining the intestine.
This causes many questions. How do luminal and nested microorganisms interact, and what is their role in our defense against pathogens and immune response regulation? Today, we do not have clear answers to many of these questions. The study of microorganisms inhabiting the mucous membrane is extremely important as they are antibiotic-resistant and can serve as the basis for regenerating of the intestinal microbiome in case of antibiotics-associated diarrhea.
The problem of bacterial drug resistance calls for special attention. If you use antibiotics without a professional’s prescription, you may be “breeding” benign bacteria, but they are resistant to similar drugs. This resistance can be shared with other pathogenic drugs through horizontal gene transfer. Uncontrolled use of antibiotics definitely impacts the speed of the distribution of pathogenic strains in the human population.
The study of human microbiota is a rapidly developing field. New data emerge with fantastic speed, and every month, we learn something new about our microscopic symbionts, which, in many ways, define our health and life itself.
References
Arumugam M., Raes J., Pelletier E. et al. Enterotypes of the human gut microbiome // Nature. 2011. V. 473. P. 174–180.
Arumugam M., Sunagawa S., Mitreva M. et al. Genomic variation landscape of the human gut microbiome // Nature. 2013. V. 493. P. 45–50.
Claesson M. J., Jeffery I. B., Conde S. et al. Gut microbiota composition correlates with diet and health in the elderly // Nature. 2012. V. 488. P. 178–184.
Dubinkina V. B., Tyakht A. V., Odintsova V. Y. et al. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease // Microbiome. 2017. V. 5(1):141.
Huttenhower C., Gevers D., Knight R. et al. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome // Nature. 2012. V. 486. P. 207–214.
Karlsson F. H., Fåk F., Nookaew I. et al. Symptomatic atherosclerosis is associated with an altered gut metagenome // Nat. Commun. 2012. V. 3:1245.
Lagier J.-C., Million M., Hugon P. et al. Human gut microbiota: repertoire and variations // Front Cell Infect. Microbiol. 2012. V. 2:136.
Ogorodova L. M., Fedosenko S. V., Popenko A. S. et al. Comparison Study of Oropharyngeal Microbiota in Case of Bronchial Asthma and Chronic Obstructive Pulmonary Disease in Different Severity Levels // Vestn. Ross. Akad. Med. Nauk. 2015. V. 6. P. 669–678.
Olekhnovich E. I., Manolov A., Samoilov A. E. et al. Shifts in the Human Gut Microbiota Structure Caused by Quadruple Helicobacter pylori Eradication Therapy // Front Microbiol. 2019. DOI: 10.3389/fmicb.2019.01902.
O’Hara A. M. & Shanahan F. The gut flora as a forgotten organ // EMBO Rep. 2006. V. 7. P. 688–693.
Petrov V. A., Saltykova I., Zhukova I. A. et al. Analysis of Gut Microbiota in Patients with Parkinson’s Disease // Bull. Exp. Bio. l Med. 2017. V. 162(6). P. 734–737.
Qin J., Li Y., Cai Z. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes // Nature. 2012. V. 490. P. 55–60.
Tyakht A. V., Kostryukova E., Popenko A. S. et al. Human gut microbiota community structures in urban and rural populations in Russia // Nat. Commun. 2013. V. 4:2469.
Tyakht A. V., Manolov A., Kanygina A. V. et al. Genetic diversity of Escherichia coli in gut microbiota of patients with Crohn’s disease discovered using metagenomic and genomic analyses // BMC Genomics. 2018. V. 19(1):968.
Wu G. D., Chen J., Hoffmann C. et al. Linking long-term dietary patterns with gut microbial enterotypes // Science. 2011. V. 334. P. 105–108.
Yatsunenko T., Rey F. E., Manary M. J. et al. Human gut microbiome viewed across age and geography // Nature. 2012. V. 486. P. 222–227.