Inhibitors of genetic reprogramming – a new page in cancer therapy
Cancer has a number of frightening features, which lie at the root of the main problems in fighting this disease. First, cancer cells are still cells of the same organism, albeit with altered behavior. Second, they can change extremely quickly, adapting to new niches when metastasizing and developing drug resistance. Third, the very mechanism underlying the emergence of cancer also differs from case to case. As a result, the disease, like a phoenix, can resurrect even after the most rigorous therapy, which ricochets at healthy cells and tissues. The author and their team have found a universal mechanism allowing the malignant tumor to survive in the high-stress environment due to activation of “dormant” genes. Moreover, chemical inhibitors of this process have already been created and are now undergoing clinical trials, offering a hope of boosting the efficiency of the existing therapies of one of the most formidable diseases of today
 This article is based on a lecture Roninson gave in May, 2019, at Sirius Educational Center in Sochi, where several major federal scientific events took place: the Third International Conference “Science of the Future” and the IV All-Russian Forum “Science of the Future – Science of the Young”.
This article is based on a lecture Roninson gave in May, 2019, at Sirius Educational Center in Sochi, where several major federal scientific events took place: the Third International Conference “Science of the Future” and the IV All-Russian Forum “Science of the Future – Science of the Young”.
The first “Science of the Future” conference was held in 2014 in St. Petersburg following an initiative from the world’s leading scientists, heads of laboratories founded as part of the Russian government scientific grants program; it was organized by the Russian Ministry of Science and Higher Education.
“Science of the Future”, which began as a conference for mega-grant holders, eventually transformed into a forum for presenting the work on modernization of the national science. In 2019, over 70 major Russian and foreign scientists took part in the conference; overall, over 600 participants gathered at Sirius center, including young scientists: senior undergraduate students, master’s degree and PhD students of the leading universities, holders of awards for outstanding young scientists, etc. This format assured close communication between different generations of scientists and exchange of ideas and experience between specialists in different research fields
At the end of the last century, when I was getting to grips with issues of cancer therapy, all cancer drugs were, as we call them now, conventional. These classical medications аffect certain molecular objects that are necessary both to normal and cancerous cells. In other words, this approach would not allow one to create a drug that would influence strictly pathological cells alone.
The mechanism underlying the action of conventional drugs can be aimed at damaging DNA either directly (the oldest cancer drugs) or by affecting topoisomerase enzymes and nucleotide synthesis, as well as at perturbation of microtubules, which make up a cell’s cytoskeleton, i. e. at universal targets, which are necessary for the normal cell cycle of any cell. The reason why these drugs work well specifically in cancer cells is that the latter cells have altered control of the cell cycle, so that they are more sensitive to disruptions of its critical stages.
Such drugs are intrinsically dangerous to the patient – everyone knows about the consequences of chemotherapy. The main challenge here is to create a drug that is efficient at a specific moment and at the safest dosage, reducing the risks to healthy cells. It is important that these drugs make the patient better as they kill pathological cells so quickly that the tumor does not always react quickly enough to launch a resistance mechanism. A good example of successful conventional therapy is testicular cancer therapy, which has a success rate of up to 90 %.
However, most of newly developed drugs nowadays are aimed at specific targets, which are required specifically by cancer cells in order to survive and support their cellular cycle. The majority of such targets were identified using methods of genomics; this is an ongoing process. Such targeted drugs are either monoclonal antibodies or “small” molecules, including inhibitors of various enzymes, growth factors, hormone antagonists, etc.
It is important that targeted drugs act on specific tumors, which carry targets for these drugs. These drugs can only be used in the context of personalized medicine, with careful selection of drugs for specific patients. Breast cancer is a good example – nowadays it is treated almost exclusively through a personalized approach.
Most of breast cancers require estrogen, a female hormone that binds to the pool of estrogen receptors (ER) inside cancer cells, boosting their proliferation. On the other hand, about 20—25 % of breast cancers are driven by HER 2 tyrosine kinase. For this reason, before starting the treatment, it is necessary to perform molecular analysis of markers to define whether the tumor cells produce estrogen receptors (or progesterone receptors, which are dependent on the former). If so, the patients are treated with targeted hormonal therapy.
If the tumor shows an amplified HER 2 gene (meaning that the gene is present in multiple copies), the patients are treated with drugs aimed specifically at that protein: antibodies (e. g. trastuzumab) or small molecule inhibitors (e. g. lapatinib). If the cancer does not belong to either of these two groups, the tumor is classified as triple negative breast cancer. Unfortunately, there are no common successful methods of treating such tumors yet.
Genomic analysis is the key to success
 Treatment strategies for all cancers are becoming more and more standard. First, the tumor is sampled for analysis to reveal various markers. Genomic methods show the presence of particular mutations; transcriptomic methods study the expression (activity) of the genes; and proteomic methods study the cellular proteins. Treatment is chosen depending on the results. In some cases, it is immunotherapy. With chemotherapy, we must determine, which features of the tumor of that particular patient allow it to thrive and grow.
Treatment strategies for all cancers are becoming more and more standard. First, the tumor is sampled for analysis to reveal various markers. Genomic methods show the presence of particular mutations; transcriptomic methods study the expression (activity) of the genes; and proteomic methods study the cellular proteins. Treatment is chosen depending on the results. In some cases, it is immunotherapy. With chemotherapy, we must determine, which features of the tumor of that particular patient allow it to thrive and grow.
Sometimes we use the available drugs, which efficiently target the tumor, or at least are approved for these purposes. In some cases, we develop new drugs. From now on, we will speak about the development of new drugs, which requires genetic analysis. I will try to explain why.
MY GRANDFATHER DIED OF CANCER I was the only child in my family. My mother and father were born in Moscow, where their parents emigrated at the beginning of the last century, having left behind their large family clans in Ukraine and Belorussia. My parents were lucky – one part of our family that was left behind was destroyed in a pogrom in 1913, and the other part – in 1941, during the Holocaust.I loved my maternal grandfather, who escaped the pogrom, dearly; when he died of cancer, I was crushed – I was only five. Because of the Russian name of the disease, рак, literally “crawfish”, I imagined a scary crustacean that sucks life out of wonderful people. It was then that I decided to devote my life to finding a cure for cancer. My mother had a Doctor of Sciences degree in tumor pathology, and I was familiar with words like rhabdomyosarcoma since I was a little child.
If not for my interest in cancer, I would have preferred to become a historian, not a biologist. I hated biology at school; to me, it was something about collecting bugs or dry leaves. However, I realized and accepted that for my purpose, biology was necessary, like a visit to a dentist may be necessary.
Like many children, I had my childhood heroes, and some of them were scientists. Among them was Lev Zilber, a legendary Soviet virologist and immunologist, who discovered in 1937 that the Siberian encephalitis virus is transmitted by ticks. For this discovery, he was “rewarded” with many years in prison camps. When working at a prison hospital, Zilber tried, whenever possible, to conduct scientific research. He discovered that some tumors in mice caught by the inmates contained an infectious agent, which had carcinogenic properties when transplanted to other animals. Zilber suspected that it was a virus, and it was found only in the smallest of the tumors. This led him to a theory that cancer can be caused by viruses, which initiate carcinogenesis but then become inactive as the tumor grows. For the 1940’s, the idea was incredibly daring and far ahead of its time.
In the early 1970’s, I entered the Biological Department of the Moscow State University, pursuing a major in Virology. My grades were good, but I knew I had no chance of finding a job at a decent research institution. The reason was simple. For instance, my young colleague, who had been appointed head of a new lab, once asked me: “Recommend me a couple of bright fellow students… unless they are Jews, of course”, without a speck of doubt that I accepted that as the natural state of things. My father, too, was fired from the symphonic orchestra where he worked for the very same reason.
This was the last straw, which persuaded our family to take a risky step upon my graduation: request permission to emigrate from the USSR. In 1979, after two tantalizing years of waiting and an extended process of migration, we ended up in the US. This is when my new life began…
In that first year in the States, having worked through the summer at the University of Maryland, I met several well-known scientists whose textbooks I had studied in Russia. They were the Nobel Prize winners D. Nathans, D. Baltimore, P. Sharp, S. Luria, W. Gilbert, and V. Ingram; the latter discovered the sickle cell anemia mutation and later became my academic adviser when I was a graduate student at MIT. Meeting these legends in person assured me that the greatest scientific breakthroughs could be made by mere mortals
Here is a good case illustrating why it is impossible to create cancer drugs outside the realm of personalized medicine. In 2004, Martha Stewart, one of the most iconic women of the nineties, a home products mogul with her own TV show, went to jail. The reason was a new cancer medication, cetuximab, a monoclonal antibody drug that affects the epidermal growth factor receptor (EGFR) tyrosine kinase transmembrane receptor, which plays a major role in stimulating tumor growth, metastatic processes, etc. ImClone, a medical research company, conducted the first clinical trials on different patient groups without individual selection. The results were ambiguous, and FDA did not approve the drug. The CEO and some of the shareholders, including Stewart, sold their stock before that decision was announced, which is a serious felony.
A few years later, a paper was published describing the action of cetuximab in colon cancer. It turned out that if the tumor cells had the K-Ras oncogenic mutation, which happens roughly in one half of all cases, the drug was completely ineffective. In other cases, it was very effective, and the results of the earlier clinical trials would have been completely different if the trials had only included patients without the K-Ras mutation. This demonstrated that genomic analysis could be the key to success – or failure – of a cancer drug.
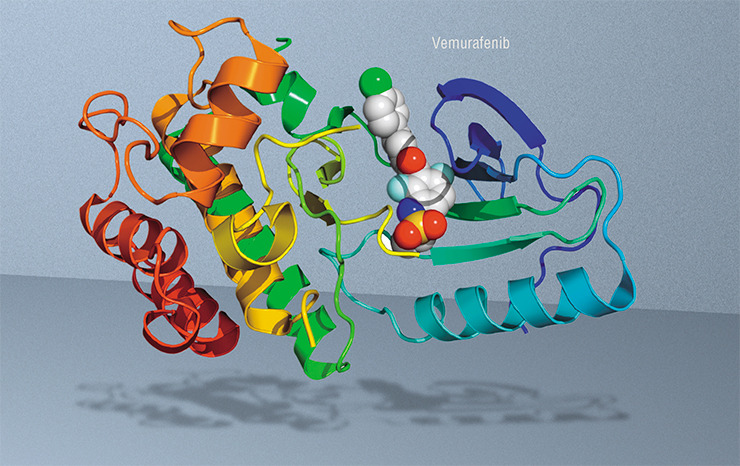
Another example deals with small molecules: nowadays, we can synthetize such molecules with very selective action on specific proteins. In our case it is the protein coded by the BRAF gene – in skin melanoma, the frequency of mutations in this gene is 30 to 70 %. The creators of a drug called vemurafenib intended it specifically for patients with this mutation.
The first results of vemurafenib therapy were striking. For example, a patient with a highly metastasized melanoma went into a nearly complete remission in the course of 15 weeks. However, the tumor eventually returned. But why? The cancer cells became resistant to the drug. Unfortunately, almost all patients develop such resistance: those treated with any targeted drugs and the majority of patients treated with conventional drugs.
What stands behind drug resistance?
How do cancer cells develop drug resistance? This phenomenon is linked to changes in the expression (activity) of genes in cells damaged by chemo-, radio-, and immunotherapy. What happens here is called reprogramming of transcription – the transfer of genetic information from DNA to the RNA molecule.
Transcriptomics gave us ways to see that in damaged cancer cells, many protein-coding genes are activated; the proteins leave the cells and stimulate growth and survival of other cancer cells, aiding the metastatic process and the formation of drug resistance. We managed to find small molecules that suppress that process by developing and applying a system of physical high throughput screening, which allowed us to evaluate over a hundred thousand various chemical compounds. At that time, the amount of work it took was enormous – nowadays we can analyze tens of millions of compounds virtually by using computer algorithms, which saves time and reduces the costs of research.
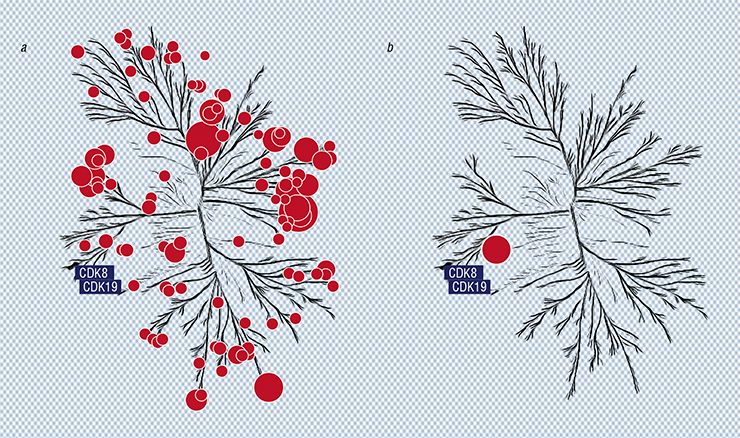
Chemists told us that structurally, our molecules looked like inhibitors of kinases – enzymes that regulate the activity and stability of a great number of proteins performing all sorts of functions, including those participating in the process of transcription. We tested our molecules on a large number of these enzymes (over 450) occurring in the human body. It turned out that our best inhibitor molecule does indeed “disarm” a great number of kinases.
Next, we began to chemically modify the “champion molecule”. At some point it turned out that its activity when suppressing transcription induced by DNA damage has improved, while some other properties disappeared. When we tested the modified molecule against the same kinase panel, it turned out that in the whole massive kinase “tree”, only two “flowers” corresponding to two kinases were inhibited (Porter et al., 2012).
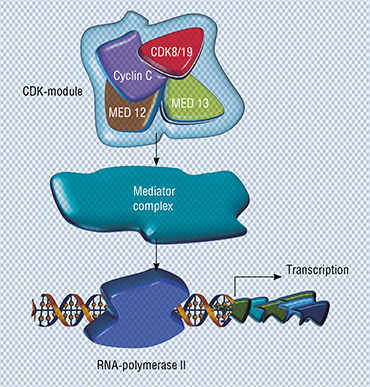 This is when our new hero hits the spotlight. Meet the CDK8 protein (cyclin-dependent kinase 8) and the closely related CDK19; both were identified using chemical genomics. It turned out that these proteins are the target we were looking for: they must be inhibited to prevent the genes associated with metastases and drug resistance from “switching on” in damaged cells.
This is when our new hero hits the spotlight. Meet the CDK8 protein (cyclin-dependent kinase 8) and the closely related CDK19; both were identified using chemical genomics. It turned out that these proteins are the target we were looking for: they must be inhibited to prevent the genes associated with metastases and drug resistance from “switching on” in damaged cells.
The CDK8 and CDK19 proteins are almost twins, identical by 80 %. Both only regulate transcription and have nothing to do with the cell cycle regulation, unlike some other members of the cyclin-dependent kinase family. However, they do interact with a number of different transcription factors, including those associated with cancer (such as the estrogen receptors mentioned above).
But if you ask me, what will happen to a cell – any cell – if you suppress CDK8/19 in it, you will not get an answer. The thing is, the effect of the inhibition is context-dependent, i. e. it depends on the specifics of the cell and its genes. As we said, these proteins only work in cooperation with other factors. All we can say is that inhibition is unlikely to seriously affect the activity of the genes which are already working. But in this manner, we can suppress the activity of those genes in the cell that were “awakened” with the help of CDK8/19.
Why does our body even need the enzymes regulating transcriptional reprogramming? To answer this question, remember when changes occur in the transcription program – it happens in the process of the development of the organism. Moreover, transcriptional reprogramming is critical to this process.
Indeed, in pregnant mice given the CDK8/19 inhibitor, embryo development came to a halt. When we gave this inhibitor to adult mice for 200 consecutive days, nothing special happened to them, neither visually, nor were there any changes in their weight, etc. But when the same drug was given to animals with breast or prostate cancer (combined with hormonal therapy) or to animals with colon cancer metastases in their liver, tumor growth slowed down drastically.
Towards clinical trials
 We tested a large number of clinical cancer drugs combined with our CDK8/19 inhibitor and found that in most cases this prevents or at least considerably slows down the development of drug resistance. There is yet one small step to take – such inhibitors need to be approved for clinical application. To achieve this, they must be effective as single agents and suppress specific cancers.
We tested a large number of clinical cancer drugs combined with our CDK8/19 inhibitor and found that in most cases this prevents or at least considerably slows down the development of drug resistance. There is yet one small step to take – such inhibitors need to be approved for clinical application. To achieve this, they must be effective as single agents and suppress specific cancers.
To find these targets, once again we turned to genomics. There is software that can predict how activation of a specific gene influences the survival of a patient with a particular type of cancer. We used one of these software products to choose an array of cancers, in which the level of CDK8/19 correlates with the worst prognosis. Among others, it included prostate cancer, cervical cancer, and esophageal adenocarcinoma. Such correlations suggest that our inhibitors can be used in these cases.
A telling example deals with breast and ovarian cancer cases. We achieve different results when we split these patients into two groups – whites and African Americans. In white people, CDK8/19 shows significant effect in breast cancer and very little effect in ovarian cancer. With African Americans, it is the opposite. This example illustrates the importance of genomic research prior to clinical trials.
Right now, we are focusing on two cancer types. The first is androgen-independent prostate cancer (androgens are male hormones). It is well known that the general approach to the treatment of prostate cancer is antihormonal therapy. As the cancer develops and progresses, the level of CDK19 rises. By the time the tumor acquires independence from androgens, the cancer becomes incurable, and we see a rise in the expression of both CDK8 and CDK19.
In experiments on laboratory mice, our CDK8/19 inhibitor combined with hormonal therapy completely suppressed the growth of tumors that had an active gene encoding the variant of androgen receptor, which made the cancer resistant to all types of available therapies. Animals did not lose weight and lived significantly longer.
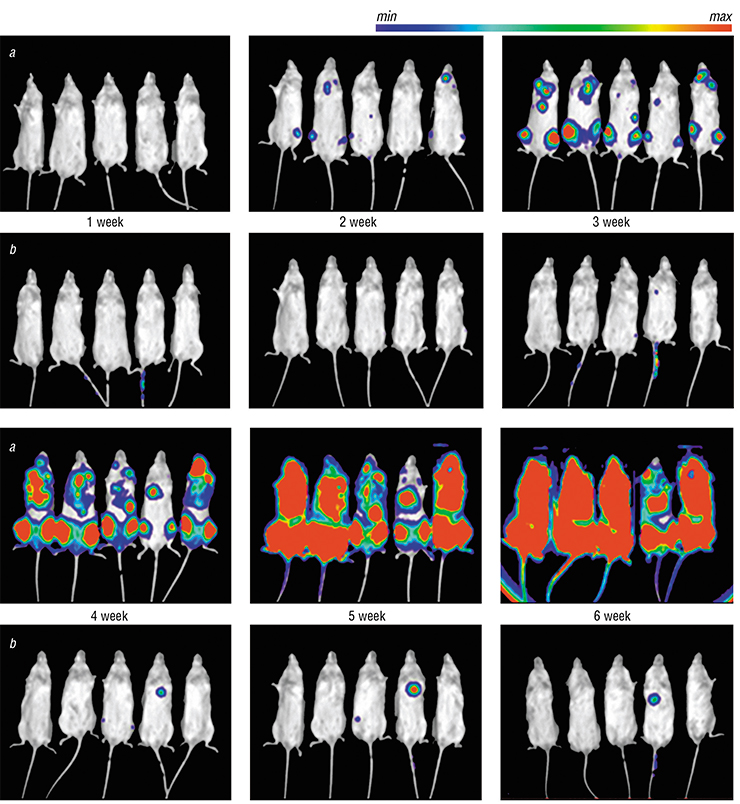
The second type of cancer is leukemia, a blood disease, and acute myeloid leukemia in particular. Currently this is the only known cancer that can be stopped by CDK8/19 inhibitors not only in animal models but in human cell culture as well. To find out how well will patients respond to our drug, we injected mice with cancer cells marked with luciferase, an enzyme, which catalyzes a chemical reaction accompanied by bioluminescence. It turned out that in control animals, the leukemia spreads through the whole body in the course of six weeks, while in animals that received the CDK8/19 inhibitor, it barely spread at all.
AGING CANCER CELLS GIVE NEW LIFE TO THE TUMOR My cancer research went through several phases. At one time, I was studying a single protein; next, it was a lot of different proteins; then a single protein again. The first one was the MDR 1 protein, which we discovered in the mid‑1980’s, it is responsible for an important mechanism of multiple drug resistance. We asked ourselves – why do tumor cells which do not normally have that protein begin to produce it actively? It turned out, when we damage these cells by certain drugs, transcription of the corresponding gene is activated in at least in some of them: DNA template guides the assembly of RNA molecules, which in turn guiding the synthesis of this protein.We discovered this phenomenon in the early 1990’s, and I gave a lot of thought to the ways of hindering this induction. Still without an answer, I switched to other things. But at one moment, we observed that chemotherapy and radiation cause a biological response within the cells, of the kind that was considered impossible at that time. I am speaking about cellular aging (senescence), a biological program that stops cell division and causes cells to change in a number of ways. It turned out that many cells that do not die after treatment with many different types of drugs enter this state.
But what does that mean for cancer cells? That cells do not divide is good – the tumor will not grow. By that time, in the late 1990’s, transcriptomics was a budding new technology, and we decided to take a look at gene expression in these “aged” cells. We found that some of the genes were suppressed, and, conversely, other genes were induced. The first group included genes associated with cell proliferation (division), which is unsurprising, because these cells did not divide. The second group was extremely interesting. Many of these genes were associated with cancer, and others encoded proteins that could affect both their own cell as well as the neighboring cells, helping the tumor survive, grow and metastasize. In addition, we found that many genes in this group were related to aging disorders (Alzheimer’s, arthritis, etc.).
This unusual response must have been caused by DNA damage, so we ran a transcriptomic analysis of the damaged cells – before they “aged”. We observed active transcriptional reprogramming in all of these cells. In the “old” cells, all these changes in gene expression were already “snap-frozen”, and they were at their maximum.
We also found that there was a central mediator of this effect on transcription – the p21 protein, which prevents cells from dividing in response to damage. If p21 alone is activated, we see the same changes in transcription as in damaged cells. The mechanism behind this was completely unclear, as no one knew back then, what that protein had to do with transcription. We only knew that it bound different cyclin-dependent kinases, which regulate cell division.
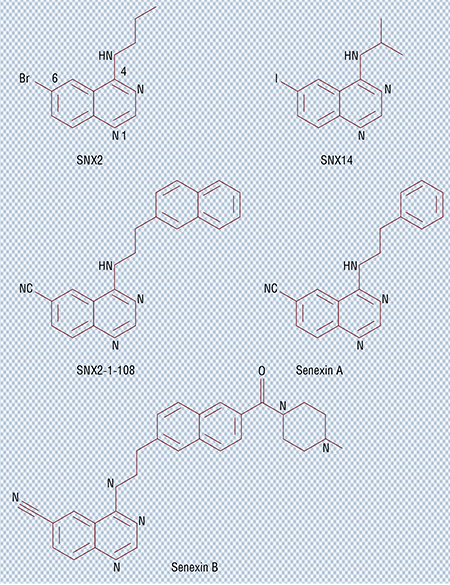
I decided that we had to find inhibitors that would stop the p21 protein from affecting transcription. This would provide a key to understanding how DNA damage in general and p21 in particular induces transcriptional reprogramming. For this purpose, I founded our company Senex. Based on p21, we developed a high-throughput screening system, which led us to CDK8/19 kinases. We discovered extremely selective inhibitors of these enzymes and have been working with them for the past 9–10 years. We also discovered that the p21 protein itself stimulates these kinases, and they in turn are responsible for transcriptional reprogramming
We tested many tumor cell samples from leukemia patients and discovered that about 40 % are highly sensitive to the CDK8/19 inhibitor. Hence, to begin treating leukemia patients, first, we need to define which patients will respond to that treatment. Using genomics and transcriptomics, we found several genes, some of which are more active in tumor cells resistant to the CDK8/19 inhibitor, and the others are more active in sensitive cells. Thus we obtained criteria to evaluate whether a specific patient will respond to our treatment. It is important to note that acute myeloid leukemia is considered an orphan disease, and the US government promotes research aimed at developing treatments for these rare disorders. This means that we can get a drug approved much faster than it takes with drugs against common diseases.
Cancer cells can become resistant to our transcriptional reprogramming inhibitors. Leukemia turned out to be the most sensitive. We performed a selection process, choosing the most drug-resistant cells, and discovered some (unpublished) things that hint at what to do in these cases. Essentially, this means we have a backup strategy.
In any case, resistance to these inhibitors develops much later than to other drugs, because the development of resistance is suppressed by the drug itself. Moreover, cancer therapy is rarely based on a single drug – there is usually a combination of several drugs. If we prove that with particular types of cancer (such as leukemia) the drug significantly improves survival, it will be quickly approved for practical application. This means that we will be able to test the drug in combination with other approved treatments. In this case, we expect an increase in the efficacy of therapy.
There is an important point: in some cases (such as colon cancer) our drug acts selectively on metastatic tumors, which is completely unique. Why? Metastases form where these cells are not supposed to grow and where they have to adapt to the stress of the “wrong” environment. They adapt specifically by transcriptional reprogramming, the “awakening” of new genes.
CDK8/19 inhibitors are developed by our American company, Senex, in close cooperation with BIOCAD, a St. Petersburg-based Russian company; the collaboration has been crucial to the progress that we have been able to achieve.

It took Senex about six years to find new molecules that suppress transcriptional reprogramming. BIOCAD has completed clinical trials of the first candidate drug, which we developed together with the Russian scientists. Next, we worked on it some more and are now planning clinical trials of the new, improved version. I hope that the moment we get our drug approved to be used in combination with other cancer treatments will breathe new life into targeted therapy.
Science First Hand asked Aleksandr Albertovich Shtil, head of the Laboratory of Cancer Cell Death Mechanisms of the N. N. Blokhin National Medical Research Center for Oncology to tell us about the problems and priorities of transcriptional reprogramming research as part of research on the molecular nature of malignant neoplasms and cancer drug development. Dr. Shtil has been working together with Dr. Roninson since the 1990’s; in 2018, they founded the Laboratory of Molecular Oncobiology at the Institute of Gene Biology of the Russian Academy of Sciences as part of a mega-grant entitled “CDK8-mediated reprogramming of gene transcription: biological mechanisms for medicine”
NEW HOPES IN ONCOLOGY There is hardly a more complicated, ambiguous and multifaceted area in medicine than oncology. Now and always, cancer has been a problem for doctors and biologists, chemists and physicists, psychologists and sociologists. Even among the gravest diseases, cancer is special; nothing sounds as serious and unsettling as cancer. Cancer research combines profound intellectual efforts and selfless actions, all-encompassing patience and hope… Indeed, the 20th century saw the retreat of deadly epidemics after the rise of antimicrobial drugs – but is there hope that the same will happen to cancer?Chemistry and biotechnology offer innumerable active compounds, which are deadly to cancer cells and more or less sparing to non-cancerous ones. At the level of cell culture, the problem is generally manageable; it gets more difficult in the body of a mouse: we must cure the experimental tumor without killing its host. Decreasing the dosage improves tolerability, but the tumor survives, too – structurally, it is too similar to normal tissue. It is easier with microbes – they do not resemble their victims at all, and devising an antimicrobial medication harmless to mammalian cells is a soluble problem. It is different in oncology… Or is it possible to find something in cancer cells that is crucial to them and yet not that important to healthy cells, and use that something to create a drug that would target cancer cells specifically instead of being universally toxic?
This targeted approach has been developing very rapidly over the past 25 years due to the incredibly fast accumulation of knowledge about the living matter. It turns out that a mechanism that exists in a noncancerous cell along with other mechanisms becomes critical to the tumor after malignant cellular transformation, and its suppression causes the tumor to die, with minimal damage to normal cells.
Gleevec is an inhibitor of Abl protein kinase, which facilitates survival of cancerous myelocytes; it is an example of a successful target-oriented strategy. Abl inhibitors have turned the tables in chronic myeloid leukemia – a fatal blood disorder. They increased lifespan and drastically improved the quality of life for patients: these drugs are well tolerated, and the pills can be taken for months and years. Molecular biology has found a special feature, which defines the biological essence of the disease – right now, this is only true for a few types of tumors.
This is only the beginning of the journey. Drugs work and patients live longer, but cancer cells struggle for survival. The more selective our tool is towards its molecular target, the more bypasses the tumor finds – and it daftly uses the biological complexity of the living matter and the functional interchangeability of its mechanisms… In these cases, we must suppress not one, but several such mechanisms, and only those, which a normal cell can do without: we cannot increase the risks of general toxicity.
Our ongoing search has led us to the most basic problems of cell function – the transcription of genes. This is where it all begins – but isn’t it the same in cancerous and non-cancerous cells? Again, molecular science opens new paths – the structurally and functionally similar CDK8 and CDK19 protein kinases are critically important in some cases and completely unimportant in other cases. Cells, in which transcription is activated in response to external stimuli, cannot do without these kinases. But not all cells, and not in all cases: again, the basic biological mechanisms work differently in different cases. Senexins – CDK8/19 inhibitors developed in Dr. Roninson’s laboratory – prevent the growth of colon cancer cells seeding in the liver, but they do not affect the growth of primary tumors – similar processes here are regulated differently. Acute myeloid leukemia and certain forms of prostate cancer are “CDK8 diseases”. Such selectivity requires personalized treatment: senexins are indicated in specific clinical cases.
Drug resistance of tumors is the problem that has accompanied Dr. Roninson throughout his scientific path, starting from the discovery of the structure of the multiple drug resistance gene. The logic of research brought him to a completely different mechanism – peculiar regulation of gene transcription at the onset of resistance in response to cancer drugs. Experiments succeeded in preventing resistance to modern drugs and magnifying their effect on tumor cells. Senexins acquire new targets, and oncology gets new hopes.
However, it is too early to celebrate: it is hardly possible to abandon the hard chemotherapy. However, targeted “reprogramming” of the disease will extends the ways to help the patients.
I am happy that Dr. Roninson’s research has reached Russia. There is a major ongoing joint project with BIOCAD and the Institute of Gene Biology of the Russian Academy of Sciences. The project is grand and polyphonic in Roninson’s special way. Like in a symphony, three independent parts unite here: developmental biology, genetics and cell physiology. The keynote is the role of the specific gene transcription mechanism and the possibilities of its suppression in cancer therapy. The conductor leads a large combined international orchestra. The audience is holding its breath…
References
Chen M., Liang J., Ji H. et al. Cdk8/19 Mediator Kinases Potentiate Induction of Transcription by Nfkappab // Proc Natl Acad Sci USA. 2017. V. 114. N. 38. P. 10208–10213.
Liang J., Chen M., Hughes D. et al. Cdk8 Selectively Promotes the Growth of Colon Cancer Metastases in the Liver by Regulating Gene Expression of Timp3 and Matrix Metalloproteinases // Cancer Res. 2018. V. 78. N. 23. P. 6594–6606.
McDermott M. S., Chumanevich A. A. Lim C. U. et al. Inhibition of CDK8 mediator kinase suppresses estrogen dependent transcription and the growth of estrogen receptor positive breast cancer // Oncotarget. 2017. V. 8(8) P. 12558–12570.
Porter D. C., Farmaki E., Altilia S. et al. Cyclin-dependent kinase 8 mediates chemotherapy-induced tumor-promoting paracrine activities // Proc Natl Acad Sci USA. 2012. V. 109. N. 34. P. 13799–13804.
Roninson I. Seeking favors from nature // Journal Cancer Biology & Therapy. 2005. V. 4. N. 7. P. 794–799.