Can we outrun what runs in the family?
Genome editing for therapy of hereditary diseases
Heredity is, in a way, the fatalism of our age. Decoding DNA sequences is akin to predicting a person’s fate. They say genes define everything: from eye color to deviant tendencies in behavior. Add hereditary diseases and mutations linked to such diseases as cancer. Can we confront the ominous doom and change our fate inscribed on the “sacred scrolls” of DNA? Yes, we can – if not tomorrow, then in the nearest future. Genetic engineering has been dealing with these problems for several decades; however, in the past few years, genome editing has been in the highlights of public attention. What has changed? The answer is in the abbreviation CRISPR/Cas
It all began in 1987 when scientists discovered strange nucleotide repeats in bacterial DNA, separated by short regions of unique sequences. A decade later, we learned that these repeating sequences, called CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) are the bacterial system of adaptive immunity, providing protection against foreign nucleic acids and bacterial viruses (bacteriophages) in particular.
But what does it have to do with hereditary diseases of humans? The answer lies in the mechanism of action of CRISPR. Bacteriophages inject bacterial cells with their genome, which is then replicated multiple times and packed into protein envelopes, producing new bacteriophages — all at the expense of the host. The bacterial defense systems, which includesurveillance and “scissor” Cas proteins, recognizes foreign nucleic acid if the host had encountered it earlier, destroys it. The intruders are defeated.
Target recognition works using the “famous” complementarity principle, which defines how pairs of nucleotides are formed in the double helix structure of DNA. The principle works in all living things on our planet, including human cells. The most important feature of CRISPR/Cas is that it is simple and universal.
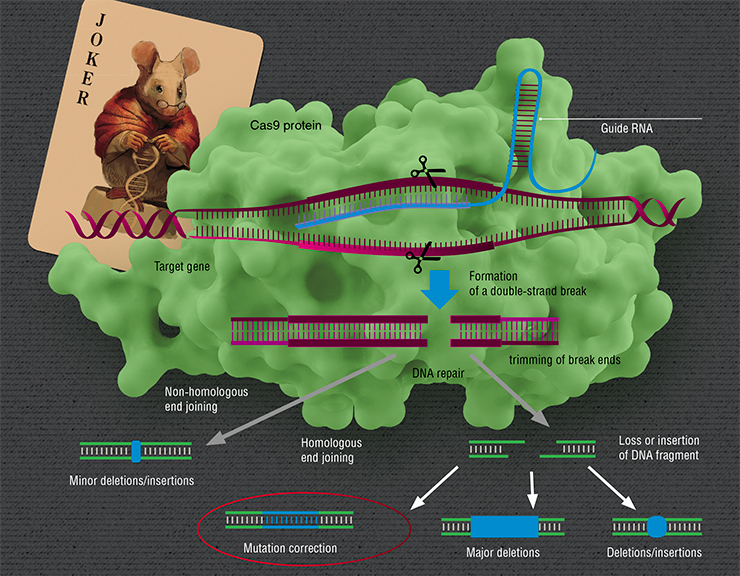
The turning point was in 2012, when Emmanuelle Charpentier, a French microbiologist, geneticist and biochemist, and Jennifer Doudna, an American biochemist and geneticist, published research demonstrating that the bacterial system CRISPR/Cas can be used to “scissor” any DNA sequence, indicating its great potential for genome editing (Jinek et al., 2012). If we know the nucleotide sequence, we can create a precisely located break in any DNA.
This gave scientists a simple and efficient tool for targeted editing of DNA in living cells, i. e. for rewriting those “scrolls”. Thousands of papers have been published since, proving that this system works in a wide variety of organisms and allows cutting any sequences of genes, including those carrying mutations causing genetic disorders (Nemudryi, 2014). In 2020, Charpentier and Doudna won the Nobel Prize in Chemistry for their discovery that revolutionized biology.
Targeted DNA repairs
So, the DNA has been snipped — what’s next? Next come repairs. Generally, breaks in DNA are not that rare: every day, in each human cell, active forms of oxygen cause about 10 thousand breaks, but the cell mends them very diligently, quickly restoring the integrity of DNA (Helbock et al., 1998). However, these breaks are random, as opposed to the action of CRISPR/Cas.
There are “therapeutic” mutations, which actually prevent the development of certain diseases. For instance, mutations in the CCR 5 gene prevent cells from being infected with HIV (Genovese et al., 2014, Liu et al., 1996), and the mutation A673T in the APP gene prevents the development of Alzheimer’s (Jonsson et al., 2012). CRISPR/Cas can be used to make the necessary changes in the genome by “breaking” the target genes or by making specific targeted changes (Cox et al., 2015)Targeted breaks made by CRISPR/Cas can be repaired in a number of ways, which differ by their mechanism, precision, etc. Depending on the reparation technique there can be different consequences. The gene can be “broken”, if the reparation causes a mutation. This effect occurs, for instance, if reparation is performed by joining non-homologous ends, a method known for its lack of precision. It can also cause a major deletion (loss of a DNA fragment) or remove a region or a whole gene.
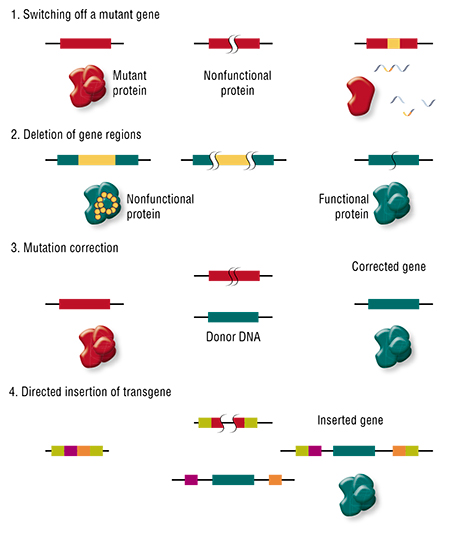 Second, a certain sequence of DNA in the cell can be “rewritten”. We know that when breaks are repaired following the homologous recombination principle, the cell uses the sister chromosome as the template to repair the damaged fragment of its DNA (our cells carry two copies of each chromosome — a maternal and a paternal one). The trick is, the cell can be “fooled” by substituting the sister chromosome with an artificially created “donor” DNA fragment. If it is similar enough to the damaged region, the cell will use it as the template.
Second, a certain sequence of DNA in the cell can be “rewritten”. We know that when breaks are repaired following the homologous recombination principle, the cell uses the sister chromosome as the template to repair the damaged fragment of its DNA (our cells carry two copies of each chromosome — a maternal and a paternal one). The trick is, the cell can be “fooled” by substituting the sister chromosome with an artificially created “donor” DNA fragment. If it is similar enough to the damaged region, the cell will use it as the template.
The cell only carries a single copy of the sister chromosome, however, we can make as many copies of the “donor” DNA as we need, which gives this artificial DNA an advantage, even if it is somewhat different from the damaged part. This way we can “fix” a mutation or insert a small fragment of new DNA.
This brings us to the therapy of genetic disorders. First, all such pathologies are different: they are caused by mutations in different genes, and these mutations can be essentially different and cause different effects in the same gene. Therefore, there are different modes of using genome editing: we can “break” a gene or simply delete the mutant region, “correct” the mutation, or, conversely, add new “therapeutic” mutations to the genome, or even a new additional transgene.
This is brilliant — in theory, but here is the problem: an adult human has tens of trillions of cells, and each of them contains the mutant gene. How can we apply this knowledge? How do we treat a real person?
Working in vitro and in vivo
In reality, there is usually no need to correct a mutation in every cell of the body. For instance, sickle cell anemia is caused by mutations in a gene coding a subunit of hemoglobin, which leads to dysfunction of only one cell type — the erythrocytes. Hereditary neurodegenerative disorders, such as lateral amyotrophic sclerosis, are caused by death of certain neuron types. This means that for treating many genetic disorders, we can target only the cells of specific organs or tissues which specifically synthesize (or not) the products of the mutant genes.
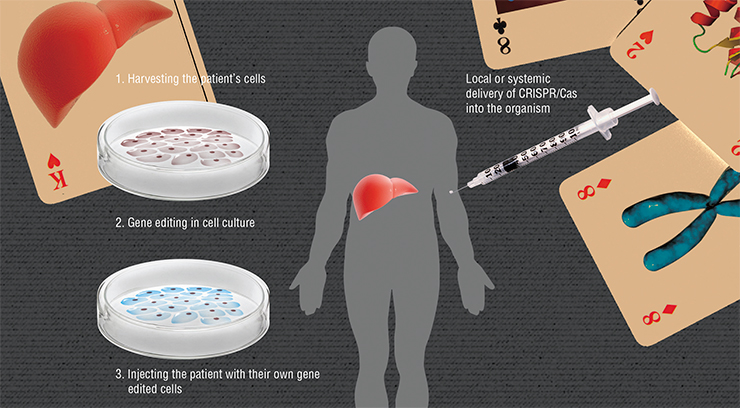
The essence of ex vivo («out of the living») genome editing consists in injecting the organism with “healthy” cells, which will, for instance, synthesize the necessary protein. However, even if we inject cells taken from a healthy donor, the recipient’s body is very likely to reject them. We must take the patient’s cells, change the mutant gene in these cells, and inject them back. This approach is especially well-developed for diseases of the blood, because harvesting and transplantation of bone marrow, which produces blood, has been done since 1959.
But what can be done in cases when such “defective” cells are difficult to harvest — for example, if the disease is manifested in the brain? Moreover, not all kinds of cells can survive in the Petri dish, away from the comfort of the organism. Another extremely promising cutting-edge technology comes to the rescue. It deals with making induced pluripotent stem cells (IPSC).
Pluripotent stem cells are immortal: in theory, they can divide indefinitely and, given certain stimuli, form cells of any tissues and organs of an adult organism. Using specific sets of stimuli, we can direct the development of stem cells into a desired cell type, e.g. neurons. This process is known as directed differentiationA relatively simple and efficient method of generating stem cells from skin cells using reprogramming was invented in 2006 by two Japanese researchers, Kazutoshi Takahashi and Shinya Yamanaka (Takahashi & Yamanaka, 2006). The technology made it possible to return virtually any cell of the body (blood, skin, fatty tissue, etc.) to the stem cell state.
The feasibility of using IPSC for cellular therapy of hereditary diseases was first demonstrated on a mouse model of sickle cell anemia (Hanna et al., 2007). Mutant human genes causing the development of this disease were inserted into the genome of laboratory mice.
Induced pluripotent stem cells were then derived from skin cells of these animals; the mutation was corrected in these cells using homologous recombination. Using directed differentiation, the researchers produced precursors of blood cells, which were then transplanted into the animals’ bodies. Not only were these cells accepted by their hosts, but they also turned into healthy erythrocytes. The treatment was a success.
Scientists have since added dozens of names to the list of hereditary diseases successfully treated with this method, and the list is still growing.
But why waste money and time on extraction and cultivation of cells if everything can be done on the spot? In theory, the versatility of CRISR/Cas allows it to work directly in the cells of a living organism. The main problem lies in efficient and safe delivery of this system to the required location.
Nowadays, CRISPR/Cas genes are often delivered into the body using viruses. The carriers of choice here are the so-called adeno-associated viruses –defective viruses which can only reproduce when accompanied by “helpers” — adenoviruses. These viruses efficiently infect human cells, but do not cause any pathologies. Viral particles with CRISPR/Cas genes in their genome are incapable of reproducing after the infection. At the same time, different serotypes of these viruses have different affinities for specific tissues: for instance, the serotype AASV prefers liver tissues, and we have the necessary technology to create artificial serotypes for any specific organ tissues.
Researchers are developing numerous nonviral delivery methods, such as packing complete molecular RNA-protein complexes into liposomes (lipid bubbles) or polymer particles. These methods are safer and allow strict dosage control.
That’s great, but…
There’s always a “but”. Researchers began to use the CRISPR/Cas technology for editing mammalian genomes less than decade ago. By now, there is a wealth of new data and extensive progress towards clinical applications; however, there are still questions to figure out for each specific genetic disorder. For instance, what is the best delivery method? Will the injected cells engraft? How safe and efficient is the treatment? Etc., etc.
Let’s take a couple of these issues and see if the scientific community has come up with any answers. One of the problems with using CRISPR/Cas is in its side effects: the system can snip regions of DNA which differ from the target region by several “letters”, or nucleotides. This carries a risk of unexpected mutations. This problem is especially relevant for in vivo genome editing when you cannot first check the results in a safe “sandbox”.
To solve this problem, three leading groups of scientists led by Jennifer Doudna, Feng Zhang and Carl June independently created mutant “improved” variants of the Cas9 protein by increasing its specificity, and the frequency of non-target effects plummeted by several orders of magnitude.
The genome editing technology can be used not only to treat hereditary diseases, but also to create “designer” babies. If we can correct a gene causing a disease, what stops us from changing eye color, lifespan, or intelligence? Chinese scientists have already used CRISPR/Cas to create beagles with a “disabled” gene coding myostatin – a factor that suppresses muscle growth, which has produced extremely muscular dogs. What keeps us from switching off this gene in a human embryo?As for the efficiency of such therapy, everything depends on the peculiarities of the disease and of the mutation causing it. There are three possibilities: correcting the mutation may improve the viability of the cells, worsen it, or there may not be any effect at all. In the first case, the corrected cells receive a competitive advantage and may eventually replace mutant cells. For instance, systemic delivery of CRISPR/Cas directly into the organism of mice with Type I tyrosinemia (a hereditary liver disease) corrects the mutation in 6 % of the liver cells. Even this small percentage of healthy cells was enough to prevent weight loss and normalize the biochemical profile of the animals’ livers. The more viable cells with the corrected mutation begin to “inhabit” the liver.
Yet if we look at hemophilia B, a corrected mutation does not improve the viability of the cells. Nevertheless, even 3—7 % liver cells producing a normal clotting factor is enough for a lasting therapeutic effect (Ohmori et al., 2017).
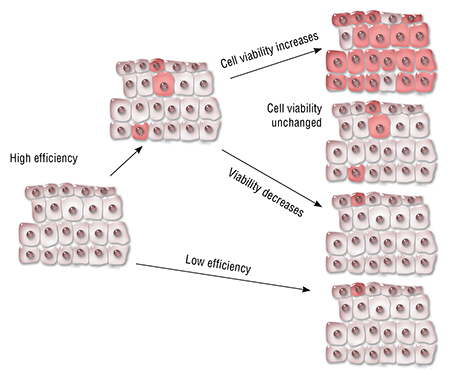 As for correcting oncogenic mutations, the viability and proliferation speed of such cells will be low compared to cancer cells, which casts reasonable doubt on the efficiency of such therapy.
As for correcting oncogenic mutations, the viability and proliferation speed of such cells will be low compared to cancer cells, which casts reasonable doubt on the efficiency of such therapy.
Nevertheless, the example of hemophilia B demonstrates that in cases when a disease is caused by a lack of an enzyme or a hormone, even a small number of cells producing this substance may be enough to make the disease milder, or even recover lost functions completely.
Various hemoglobinopathies and Duchenne muscular dystrophy remain the classic model pathologies in genome editing research. It is for these diseases that scientists were able to prove the feasibility of such treatment and show that cells with the corrected mutation demonstrate a healthy phenotype. In the case of Duchenne’s, not only did such cells successfully engraft themselves into the muscle tissue of adult mice, but they also improved the functional values of the whole muscle.
The CRISPR/Cas9 systems opens up new horizons for humankind, yet one must remember that it is not a magic wand, but just a tool. And we must learn to use this tool for each task.
The major step — which has already been taken — is taking this technology out of the lab. There are companies that are implementing the CRISPR/Cas technology both for medical and non-medical purposes. For instance, it is used to produce modified microbes for biotechnology as well as modified plants.
We can expect clinical applications of the new technology within a decade. In 2016, the first clinical trials of a new immunotherapy method for metastatic non-small cell lung cancer using “edited” T-lymphocytes began in China. In October of 2020 CRISPR Therapeutics (co-founded by Emannuele Charpentier in 2013) reported successful use of gene editing for treating β-thalassemia and sickle cell disease in humans. A year after receiving treatment all seven enrolled patients with β-thalassemia were transfusion-independent, while three patients with sickle cell disease had no vaso-occlusive crises (Frangoul et al, 2020). These new kindles hope that these treatments will be available in nearest future and more has yet to come.
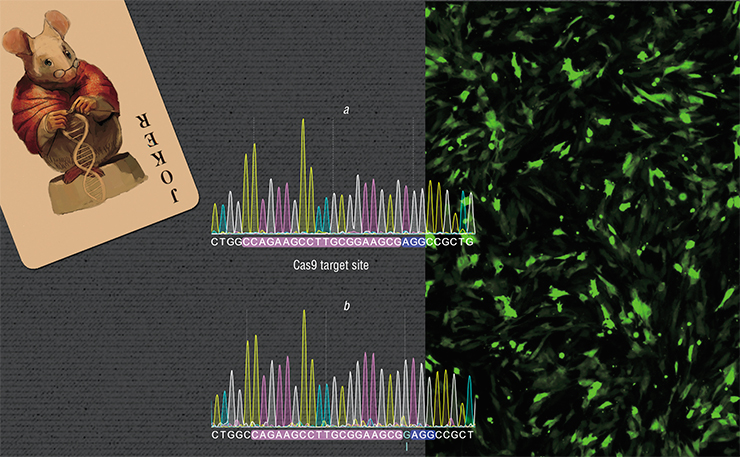
The research is carried out in Brattleboro rats – a strain of laboratory rats representing a model of a genetically determined disease, which manifests in the deficiency of a hormone called arginine vasopressin. As the result, the animals develop diabetes insipidus with the characteristic excessive thirst and diuresis. We created a strain of cells in which the mutation in the gene coding this hormone has been corrected.
In this case, the task was complicated by the fact that the nucleotide sequence of the vasopressin gene is very similar to the gene coding another hormone – oxytocin. Moreover, in the region where this mutation occurs in Brattleboro rats, these genes are almost identical. Nevertheless, we successfully applied CRISPR/Cas in the vasopressin gene without non-target “snips” of DNA in the oxytocin gene. The next step in the research is to inject the corrected cells into the animals and evaluate the therapeutic effect
The possibilities provided by the CRISPR/Cas technology are both exciting and frightening. First Chinese scientists, and later their colleagues from the United States succeeded in editing human embryos. A group of American scientists led by Shoukhrat Mitalipov “corrected” a mutation in a human embryo that causes hypertrophic cardiomyopathy. These embryos were obtained using artificial insemination by using a healthy egg and sperm from a carrier of the mutation. Following the current ethical standards, the experiment was terminated when the embryos were at the blastocyst stage. However, with the already available reproductive technology these embryos could have been implanted to surrogate mothers. In the late 2018 world was shaken when He Jiankui, researcher from China, announced that two gene-edited twin girls were born. Group led by Jiankui edited the CCR 5 gene in the embryos to introduce a mutation that confers resistance to HIV. This scandalous work was widely condemned and led to investigation resulting in imprisonment of He Jiankui.
This brings up a serious ethical issue: do we have the right to change human DNA? Or, inversely, how ethical is inaction if it dooms the child to a life of suffering? When is the use of gene editing justified?
Genetic changes inserted into embryos will persist in all cells of the adult — and they will be passed on to the future progeny. What sort of impact will the distribution of such modified genes have on the human population, evolution-wise? That is, even if we disregard the risk of the emergence of new eugenics movements in the society?
This is why Doudna, a colossal authority in the scientific world, is appealing to her colleagues not to rush with using this technology on human embryos until there are internationally accepted ethical and legal guidelines to regulate it. Nowadays, the future of CRISPR/Cas is discussed all over the world not only at scientific conferences, symposiums, and meetings, but also with the general public. The decisions made may be crucial to the future of humanity and the world.
Such international meetings on modern genome editing technology are also regularly held in Novosibirsk Akademgorodok: the took place in the September of 2018, and more are planned. Among other things, the guests discuss CRISPR/Cas and its future in the Russian Federation.
References
Cox D. B., Platt R. J., Zhang F. Therapeutic genome editing: prospects and challenges // Nat Med. 2015. V. 21. N. 2. P. 121—131.
Frangoul H., Altshuler D., Cappellini D., et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia // New England Journal of Medicine. 2020.
Jinek M., Chylinski K., Fonfara I. et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity // Science. 2012. V. 337. N. 6096. P. 816—821.
Ma H., Marti-Gutierrez N., Park S. W. et al. Correction of a pathogenic gene mutation in human embryos // Nature. 2017. V. 548. N. 7668. P. 413—419.
Nemudryy, A. A., Valetdinova, K. R., Medvedev, S. P., Zakian, S. M. TALEN and CRISPR/Cas genome editing systems: Tools of discovery // Acta Naturae, 2014, V. 6 (22), 2014, P. 19-40.







