Viruses and Bacteria: The Great Confrontation
The modern genome editing technology, which has been used successfully on various animals, plants, fungi, and bacteria, is based on research about CRISPR-Cas bacterial systems. Originally, they were thought to participate in DNA repair, but in 2007 scientists found out the real purpose of these systems: they combat bacterial viruses, i. e., bacteriophages. It took scientists as little as nine years to make a giant leap from understanding the mechanism of bacterial immunity to human genome editing, and now they are making the first experiments on editing the DNA of human embryos. Moreover, bacteria have other immune mechanisms too, and studying them might lead to new breakthroughs in biomedicine
Bacteriophages are viruses that exclusively attack bacteria. During infection, they take under control all the life processes of the bacterial cell, virtually transforming it into a factory to produce viral progeny. Eventually, the cell dies, and the new viral particles come out to infect new bacteria.
Although natural phages are very abundant and highly diverse, we rarely encounter them face-to-face. However, there are situations when the activity of these viruses does not go unnoticed. For example, producers of cheeses, yoghurts, and other lactic acid products often have to deal with viral attacks on milk-fermenting bacteria. In most of these cases, the phage infection spreads fast and “good” bacteria die, leading to significant economic losses (Neve et al., 1994).
To address this challenge of the dairy industry, scientists have obtained bacteriophage-resistant strains of lactic acid bacteria. It was these applied studies that revealed the specific mechanisms of how bacteria can avoid infection. Simultaneously, this research revealed the ways of how viruses overcome bacterial defenses (Moineau et al., 1993).
Foreguarded is forearmed
As of today, we know five major—quite cunning—defense mechanisms that bacteria have developed in their incessant struggle against viruses: changing the cell membrane receptor; superinfection exclusion; the abortive infection systems; the restriction–modification systems; and, finally, the CRISPR-Cas systems.
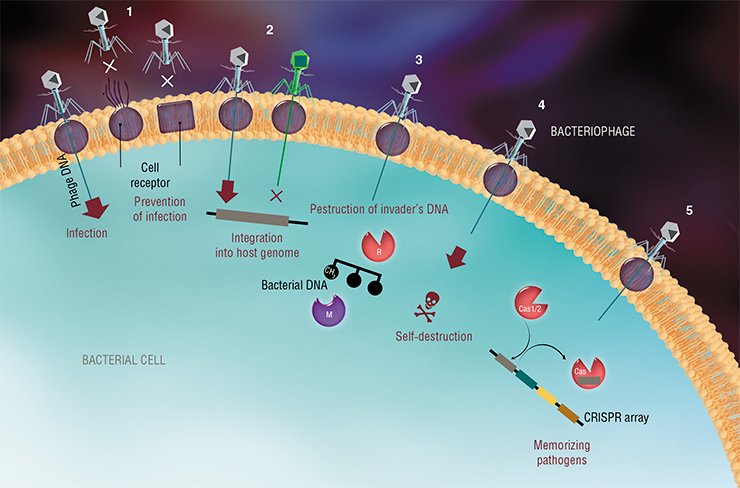
A viral attack begins when a phage attaches through a specific receptor on the surface of a bacterial cell. However, if the receptor is lost or its structure is changed, there is no viral binding. Bacteria can change their receptors depending on environmental conditions such as density and diversity of microorganisms in the media and availability of nutrients (Bikard et al., 2012). An interesting example is bacteria of the species Vibrio anguillarum, which can create a biofilm, i. e., a dense cell layer attached to a surface. These bacteria have quorum sensing feature: when the density of the cells increases, they lower the production of the receptor the virus can bind. Thus, the biofilm becomes almost totally resistant to infection (Tan et al., 2015).
However, the bacterium may suffer from losing its receptors since they perform a variety of important functions, e. g., the transport of nutrients or the formation of intercellular contacts (Lopez-Pascua et al., 2008). Thus, each bacterium–bacteriophage pair has found, in the course of the evolution, an optimal solution ensuring an acceptable level of protection while preserving the possibility of bacterial growth in various environmental conditions.
The next protective mechanism is superinfection exclusion. It is known that bacteriophages can infect bacteria in two main ways: the lytic way, which leads to the rapid death of the infected bacterium with a release of viral progeny, and the lysogenic way, when the viral genetic material integrates into the bacterium’s genome and replicates with the host DNA, without causing any damage to the cell. When a cell becomes a lysogen, infection with any other phage is unwanted for the intracellular virus (prophage).
Indeed, many viruses that have embedded their DNA into the cell genome will restrain a newly arrived bacteriophage (superinfection) by means of special repressor proteins that do not allow the invader’s genes to work (Calendar, 2006). Moreover, some phages even prevent other viral particles from penetrating into the infected cell by affecting its receptors. As a result, a bacterium infected with the virus has an obvious advantage over its uninfected siblings.
In 1978, the discovery of restriction enzymes was awarded with the Nobel Prize, which went to a Swiss geneticist Werner Arber and American microbiologists Daniel Nathans and Hamilton Smith. Research into the restriction–modification systems led to the development of molecular cloning technology, which is now widely used throughout the world. Restriction enzymes help cut out genes from the genome of one organism and insert them into that of another organism to obtain chimeric recombinant DNA that does not exist in nature. Scientists use various modifications of this approach to isolate and investigate individual genes. In addition, this enzyme is widely used in pharmaceutics, e. g., for the production of insulin or therapeutic antibodies: all such drugs have been developed by molecular cloning, i. e., are a product of gene modificationDuring infection, all the resources of the bacterial cell are channeled into the production of new viral particles. If any vulnerable bacteria happen to be near such a cell, the infection will spread quickly and kill most of them. However, for such cases, the bacterium has the so-called abortive infection systems, which guide it to a programmed death. Of course, this altruistic mechanism will not save the infected cell itself, but it will stop the spread of the viral infection for the benefit of the entire population. The abortive infection systems in bacteria are very diverse, but the details of their functioning have not been adequately studied yet.
Another type of antiviral defenses in bacteria is the restriction–modification systems, which include genes coding two enzyme proteins—restriction enzyme and methylase. Restriction enzyme recognizes certain DNA sequences 4–6 nucleotides long and makes double-strand breaks in them. Methylase, on the contrary, covalently modifies these sequences by adding methyl groups to individual nucleotide bases to make them unrecognizable for the restriction enzyme.
All the DNA sites in a bacterium containing such a system are modified. Then, if the bacterium is infected with a virus whose DNA contains no such modification, restriction enzyme will protect it from infection by destroying the viral DNA. Many viruses combat the restriction–modification systems by not using genome sequences recognizable by restriction enzyme; obviously, virus variants with a different strategy have not left any offspring.
The last and currently most fascinating system associated with bacterial immunity is the CRISPR-Cas system, which allows bacteria to record information about the phages they have encountered in their life into their genome and pass it on to the daughter cells. These memories make it possible to recognize phages DNA and to resist it more effectively in the case of repeated infections. The CRISPR-Cas systems are currently in focus of research as they are the core of the revolutionary genome editing technology, which might help future generations to treat genetic diseases and create new breeds and varieties of agricultural animals and plants.
Know your enemy
The CRISPR-Cas systems are a unique example of adaptive immunity in bacteria. When a phage injects DNA into the cell, Cas proteins incorporate viral DNA fragments of 25—40 nucleotides in lenght into a specific region of the bacterial genome (Barrangou et al., 2007). These fragments are called spacers; the chromosome region where spacers are incorporated is called a CRISPR array (Clustered Regularly Interspaced Short Palindromic Repeats); and the very process of acquisition of spacers is called adaptation.
For a cell to use spacers in its fight against phage infection, there is another process controlled by Cas proteins, the so-called interference. Its idea is as follows: the CRISPR array transcription creates a long RNA molecule, which is cut by Cas proteins into short sequences called protective CRISPR RNAs (crRNAs), each containing one spacer. The Cas proteins together with the crRNA molecule form an effector complex, which scans the cell’s entire DNA for sequences identical to a given spacer (protospacers). When the effector complex finds protospacers, the Cas proteins cleave them (Westra et al., 2012; Jinek et al., 2012).
CRISPR-Cas systems have been found in most prokaryotes—bacteria and archaea. All the known CRISPR-Cas systems work on the same principle; however, the details of their individual mechanisms may differ considerably. The greatest differences are associated with the structure and functioning of the effector complex, which is why researchers distinguish between several types of CRISPR-Cas systems. As of today, the literature contains description of six types of unrelated CRISPR-Cas systems (Makarova et al., 2015; Shmakov et al., 2015).
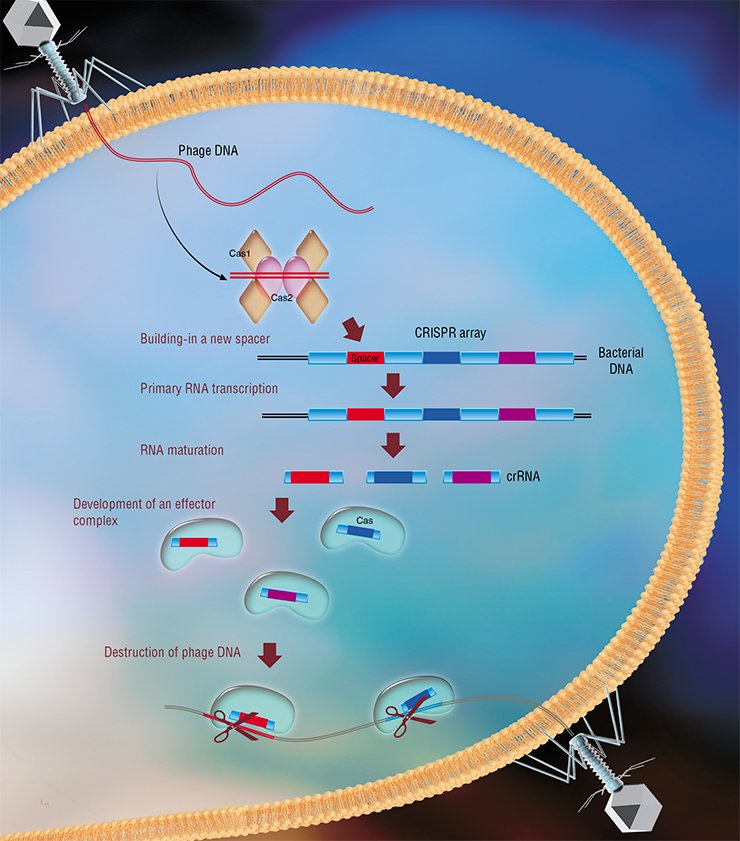
The most studied system is the type I CRISPR-Cas system, which is found, e. g., in the bacterium Escherichia coli, a favorite object of molecular biology research. The effector complex in this system consists of several small Cas proteins, each of which is responsible for different functions: cutting the long non-coding crRNA, binding short crRNAs, and searching for and then cutting the target DNA.
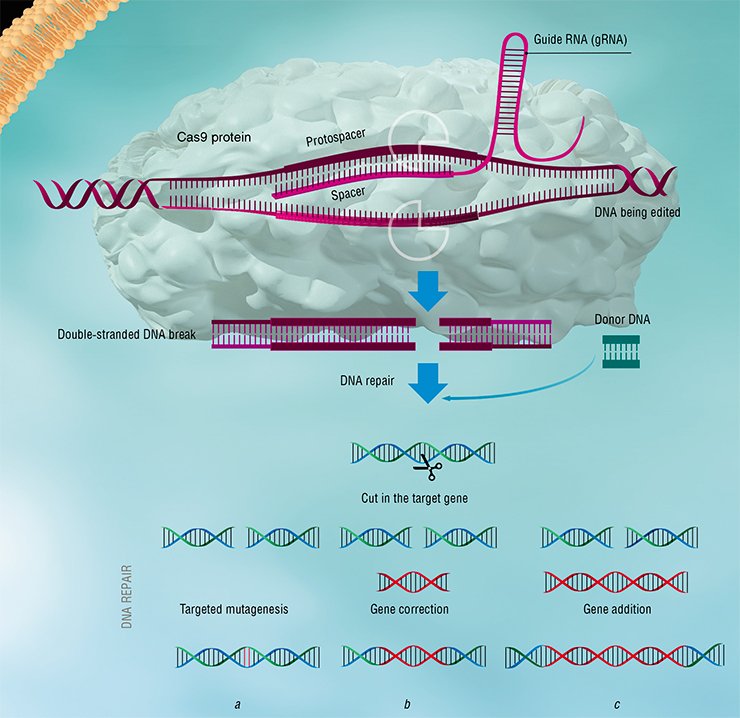
The effector complex in the type II systems is formed by one big protein Cas9, which can do all the work on its own. It is the simplicity and relative compactness of these systems that guided the development of DNA editing technology. This is a method whereby the bacterial protein Cas9 and crRNA, which is called guide RNA (gRNA), are delivered into eukaryote cells (e. g., in humans). This gRNA contains, in place of a viral spacer, a target sequence consistent with a genome region that is of interest for research, e. g., where there is a disease-provoking mutation. Today, it is quite easy to obtain gRNA that would suit any taste.
The Cas9–gRNA effector complex makes a double-strand break in a DNA sequence matching the guide RNA. If we incorporate into the cell, together with Cas9 and gRNA, a DNA sequence without mutation, the broken region will be restored from the matrix of the correct copy! Thus, we can use different gRNAs to correct unwanted mutations and make directed changes in target genes. The Cas9–gRNA complex can be programmed to recognize targets with a very high accuracy, and this method is, on the whole, so simple that it triggered an exponential growth of research on genome editing of plants and animals (Jiang & Marraffini, 2015).
Arms race
In the course of evolution, bacteria and bacteriophages developed a range of “arms” that either give them an advantage in fighting their enemy or an ability to evade the enemy’s attack.
Being an environmental factor, bacteriophages induce targeted changes in bacterial genome, which are inheritable and give bacteria a distinct advantage by protecting them against repeated infection. Therefore, we can consider the CRISPR-Cas systems as an example of Lamarckian evolution, whereby acquired characteristics are inheritable (Koonin et al., 2009)Speaking about CRISPR-Cas systems, if a phage develops a mutation in the protospacer, the effector complex will be less effective in recognizing the phage, giving it an opportunity to infect the cell. However, the bacterium will not ignore the attempt to circumvent CRISPR-Cas either: it will respond with a dramatic increase in the efficiency of acquisition of new spacers from the DNA of the already familiar, albeit mutated, phage. This phenomenon, called primed adaptation, multiplies the protective effect of CRISPR-Cas systems (Datsenko et al., 2012).
Some bacteriophages respond to CRISPR-Cas systems in a bacterial cell by producing specific anti-CRISPR proteins that can bind with Cas proteins and block their functions (Bondy-Denomy et al., 2015). Another contrivance is to replace the viral genome regions targeted by the CRISPR-Cas system by genome regions of related viruses with a different nucleotide sequence (Paez-Espino et al., 2015).
The results obtained by our laboratory show that infected cells do die even when they have CRISPR-Cas protection, but they limit the quantity of viral progeny. Therefore, it would be more correct to consider CRISPR-Cas as abortive infection systems rather than real immune systems.
Given the ongoing advancement of bioinformatic search algorithms, coupled with the expansion of research scope to include more prokaryotic genomes into the analysis, it is reasonable to expect the discovery of new types of CRISPR-Cas systems in the near future. Another task is to find out the detailed mechanisms underlying the operation of many recently discovered systems. For example, an article published in Science in 2016 on the analysis of the type VI CRISPR-Cas system describes a C2c2 protein forming an effector complex with crRNA in order to degrade RNA rather than DNA (Abudayyeh et al., 2016). In the future, such an unusual property may be used in medicine to regulate the activity of genes by changing the number of RNAs encoded by them.
Studies of the strategies used by bacteria in their struggle against bacteriophages might seem too theoretical and remote from the challenges of practical medicine. However, this research has brought invaluable benefits to mankind, as evidenced by the methods of molecular cloning and genome editing, i. e., directed introduction or removal of mutations and changes in the level of transcription of certain genes.
The rapid advancement of molecular biology methods allowed the development, only a few years after the discovery of the CRISPR-Cas action mechanism, of a working genome editing technology that is able to fight diseases that were previously deemed incurable. Widely available and simple, this technology might serve as a basis for human and veterinary medicine, agriculture, and biotechnology in the future, which will broadly apply directed and safe gene modifications.
There is no doubt that further research exploring the interactions between bacteria and their viruses will open new vistas that we now cannot even imagine.
References
Abudayyeh O. O., Gootenberg J. S., Konermann S. et al. C 2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector // Science. 2016. V. 353: aaf5573.
Barrangou R., Fremaux C., Deveau H. et al. CRISPR provides acquired resistance against viruses in prokaryotes // Science. 2007. V. 315. P. 1709–1712.
Bikard D., Marraffini L. A. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages // Curr. Opin. Immunol. 2012. V. 1. P. 15–20.
Bondy-Denomy J., Garcia B., Strum S. et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins // Nature. 2015. V. 526. P. 136–139.
Calendar R., Abedon S. T. The Bacteriophages // 2nd Ed., Oxford University Press. 2006.
Datsenko K. A., Pougach K., Tikhonov A. et al. Molecular memory of prior infections activates the CRISPR-Cas adaptive bacterial immunity system // Nat. Commun. 2012. V. 3. P. 945
Jiang W., Marraffini L. A. CRISPR-Cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems // Annu. Rev. Microbiol. 2015. V. 69. P. 209–28.
Jinek M., Chylinski K., Fonfara I., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity // Science. 2012. V. 337. P. 816–821.
Koonin E. V., Wolf Y. I. Is evolution Darwinian or/and Lamarckian? // Biol. Direct. 2009. V. 4. P. 42.
Lopez-Pascua L., Buckling A. Increasing productivity accelerates host-parasite coevolution // J. Evol. Biol. 2008. V. 3. P. 853–860.
Makarova K. S., Wolf Y. I., et al. An updated evolutionary classification of CRISPR-Cas systems // Nat. Rev. Microbiol. 2015. V. 11. P. 722–736.
Moineau, S., Pandian S., Klaenhammer T. R. Restriction/modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the dairy industry // Appl. Envir. Microbiol. 1993. V. 59. P. 197–202.
Neve H., Kemper U., et al. Monitoring and characterization of lactococcal bacteriophage in a dairy plant // Kiel. Milckwirtsch. Forschungsber. 1994. V. 46. P. 167–178.
Nuñez J. K., Harrington L. B., et al. Foreign DNA capture during CRISPR-Cas adaptive immunity // Nature. 2015a. V. 527. P. 535–538.
Nuñez J. K., Kranzusch P. J., et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity // Nat. Struct. Mol. Biol. 2014. V. 21. P. 528–534.
Nuñez J. K., Lee A. S., Engelman A., Doudna J. A. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity // Nature. 2015b. V. 519. P. 193–198.
Paez-Espino D., Sharon I., et al. CRISPR Immunity Drives Rapid Phage Genome Evolution in Streptococcus thermophilus // MBio. 2015. V. 6: e00262–15.
Shmakov S., Abudayyeh O. O., Makarova K. S., et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. // Mol. Cell. 2015. V. 60. P. 385–397
Tan D., Svenningsen S. L., Middelboe M. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. // mBio 2015. V. 6: e00627–15.
Westra E. R., van Erp P. B., Künne T., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3 // Mol. Cell. 2012. V. 46. P. 595–605.
This work was supported by the RFBR (no. 16-34-01176)











