Oxylipins: Evolution of Biochemical “Esperanto”
Every time we spill hot tea on ourselves, hurt our knees on a pavement, or get infected with flu, our damaged tissues start synthesizing bioactive substances – oxylipins. They are the first distress signal and a ‘cry for help’ from injured cells. All higher multicellular organisms – animals, plants, fungi, and algae – use strikingly similar compounds for their cell-to-cell communication. Some unicellular organisms – bacteria and protozoa – can also synthesize this kind of compounds; however, it was not clear until recently what their purpose is. Bioinformatic methods have helped to shed light on this question and concurrently identify a new target to combat antibiotic resistant bacteria
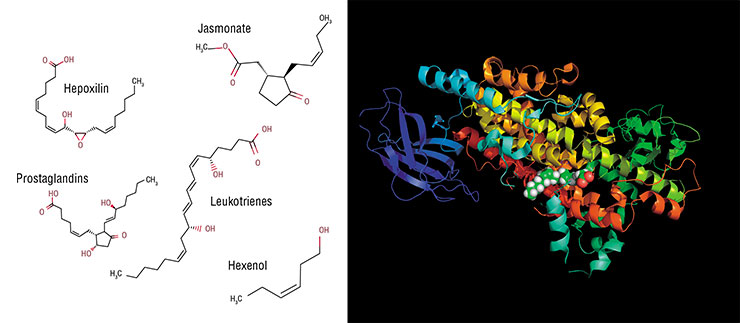
Human oxylipins are a group of bioactive molecules that orchestrate inflammation, a universal defense response of an animal organism. They regulate almost all components of this process: movement of leukocytes towards an injury site, leukocyte activation, increase in vascular permeability, blood clotting, etc. Notably, human oxylipins encompass both pro-inflammatory (e. g., leukotrienes and the majority of prostaglandins) and anti-inflammatory compounds (e. g., lipoxins).
This function of oxylipins is familiar to everyone who has ever taken a tablet of aspirin or ibuprofen to alleviate inflammation – these drugs target and block prostaglandin biosynthesis. Drugs blocking leukotriene biosynthesis or leukotriene receptors (e. g., zileuton or montelukast) are also used in medicine – for treating asthma.
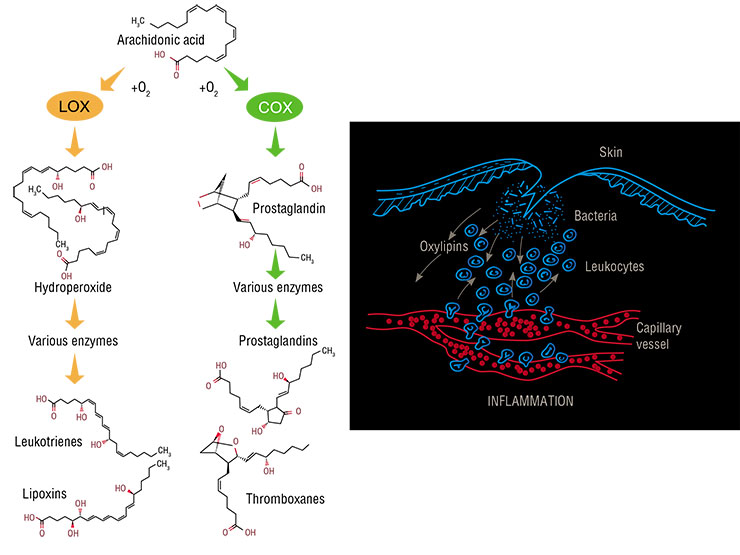
All oxylipins are derived from polyunsaturated fatty acids having several double bonds (C=C) in a carbon ‘tail’. The abundance of these bonds makes these fatty acids prone to oxygenation, which results in derivatives with an –OH or =O group; it is these derivatives that are called oxylipins. Of all the fatty acids, only arachidonic acid, with its four isolated double bonds, is used by human cells for oxylipin biosynthesis. Since its molecule contains 20 carbon atoms, all animal and human oxylipins are called eicosanoids (from the Ancient Greek word for ‘twenty’).
There is only a small amount of free arachidonic acid in the cell cytoplasm; therefore, upon injury, a special enzyme splits membrane phospholipids and literally tears this acid out from the membrane. Its further oxidation is performed by two enzymes: cyclooxygenase and lipoxygenase. The two enzymes catalyze essentially the same lipid peroxidation reaction, but they do it in different places, thus yielding products of different configurations. For instance, cyclooxygenase forms a pentatomic ring in the middle of a molecule.

There are a total of six types of lipoxygenases that participate in prostaglandin and leukotriene production. Firstly, they transform arachidonic acid into quite unstable hydroperoxides, which in turn reorganize spontaneously into epoxides, and then the whole enzyme set brushes them up and converts them into final products. There are also two types of cyclooxygenases that synthesize prostaglandins.
All the above facts are familiar to biomedical specialists. It is intriguing, however, that compounds strikingly similar to oxylipins also occur in other kingdoms of life.
Plants: «Help me, I’m being eaten!»
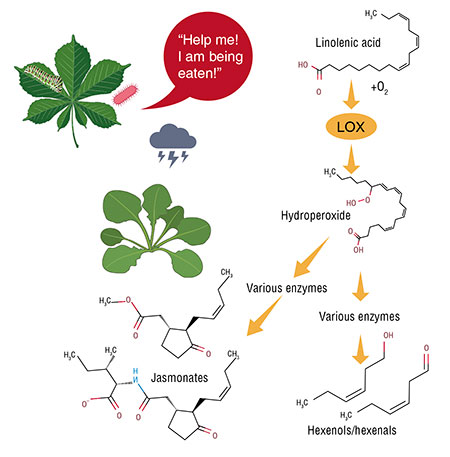
Plant cell membranes contain no arachidonic acid, but they contain instead a similar linoleic acid with 18 carbon atoms and 3 isolated double bonds (de León, Hamberg, Castresana, 2015). Plants use this fatty acid for biochemical transformations strikingly similar to the biosynthesis of our eicosanoids. The resulting metabolites are called octadecanoids, or, for clarity, simply oxylipins.
The initial step in producing these compounds is similar to that occurring in animals. Plants have their own lipoxygenases, i. e., animal enzymes homologues synthesizing hydroperoxides from linoleic acid. The next step is where the differences begin. Two enzymes readily catch a part of hydroperoxide molecules and “cut” them into volatile alcohols and aldehydes (we cannot do so as all our oxylipins are non-volatile).
The rest of hydroperoxide is processed by a series of enzymes bearing no resemblance to human ones (Heldt, 2011). As a result, jasmonic acid arises, which is called together with its derivatives (esters, glycosides etc.) jasmonates. In terms of their chemical structure, they could be considered analogues of our prostaglandins, but plants synthesize them in a different way.

In any case, there is an obvious biochemical similarity between our oxylipins and plant oxylipins. But what about their functions? At first thought, the idea of their similarity seems wacky because plants have no inflammation. In contrast to their animal counterparts, plant cells are immured in fast cellulose walls, so they are completely incapable of rushing to the rescue of cells in distress, as our leukocytes do.
On the other hand, the functional similarity appears to be amazing in the larger scheme of things. After all, “inflammation” means a stereotypic injury response that is characteristic of animals. But plants also do respond to injuries, albeit in a different way. And their injury reactions are coordinated with plant-type oxylipins.
In plants, as in animals, oxylipins regulate the cellular response to the damage caused by injury or pathogen invasion. Instead of inflammation, they trigger the release of a wide range of defence chemicals, i.e., phytoalexinsWhen a pathogen invades, a plant brings real “chemical weapons” into play. Plants run rings around us in terms of the secondary metabolite synthesizing ability (secondary metabolites are substances that are not involved in a plant’s own metabolism). For instance, invading bacteria or fungi may face phytoalexins (small-molecule antibiotics), defensins (small antimicrobial peptides similar to human counterparts), and a set of enzymes capable of destroying the cell wall of an uninvited guest (Freeman, Beattie, 2008; Heldt, 2011). So, plants do have immunity; it is only very dissimilar to ours.
 Jasmonates, as well as volatile aldehydes and alcohols (which are synthesized together with them), trigger an immune response cascade in a plant organism. This is a pivotal similarity to vertebrate eicosanoids – both plant and animal oxylipins are involved in immune response regulation.
Jasmonates, as well as volatile aldehydes and alcohols (which are synthesized together with them), trigger an immune response cascade in a plant organism. This is a pivotal similarity to vertebrate eicosanoids – both plant and animal oxylipins are involved in immune response regulation.
At the same time, these compounds act through different receptors in these two kingdoms, which seems amazing. In animals, they predominantly activate serpentine-like membrane-bound G-protein-coupled receptors. Plants have no homologues of these oxylipin receptors; besides, their oxylipin receptors are poorly elucidated. It is only known that their receptor for jasmonates is a multi-protein complex which has no resemblance to our oxylipin receptors (Mach, 2009).
Oxylipins play a bigger role in plants than in animals – in plants, they are also a part of the stress response in general. These signal molecules orchestrate a plant’s response to drought, excessive salinity or light, or other environmental challenges. While our prostaglandins are just inflammation mediators, their plant counterparts – jasmonates – are actual stress hormones!
Essentially, oxylipins switch plants from a “growth mode” to a “defense mode”. In particular, they inhibit photosynthesis and cell division, thus slowing down all the biological functions of a plant in the beginning of the leaf fall before a long winter (Savchenko, Zastrizhnaya, 2014).
Oxylipins help plants to overcome yet another challenge, i. e., the inability to communicate verbally or nonverbally. Animals use sounds, movements, or words (in the case of humans) for communication with their conspecifics, but plants cannot do so. But there is chemistry between them.

Anyone who has ever smelled the sweet scent of freshly mown grass knows what plant “crying” is – the fragrance of volatile oxylipins (aldehydes and alcohols) deriving from fatty acids when a cell is injured. Plants respond to herbivory attacks in the same way.
Plants “cry” for a good reason. For instance, one of volatile jasmonates – methyl jasmonate – is able to pass from one plant to another by air and initiate neighbors’ defense reactions in advance (Jang et al., 2014). Moreover, plants use this way of communication to call other species for help, e. g., parasitoid wasps, a natural enemy of leaf-eating larvae (Heldt, 2011).
The functions of plant oxylipins extend beyond communication – some of them are antimicrobial agents and are themselves able to kill bacteria and fungi. But the communication function seems to be especially important given the ecological conditions set for plants by evolution. In these terms, the plant and animal oxylipin signalling systems seem to be very similar because both animal and plant cells cry with these signals: “Help me, I am being eaten!”
Algae and fungi: “I like you!” but “Don’t push!”
Here, we come on shaky grounds because oxylipins and their roles in other organisms are not so well understood. For example, our knowledge on this topic regarding unicellular red, brown, and diatom algae is quite piecewise.
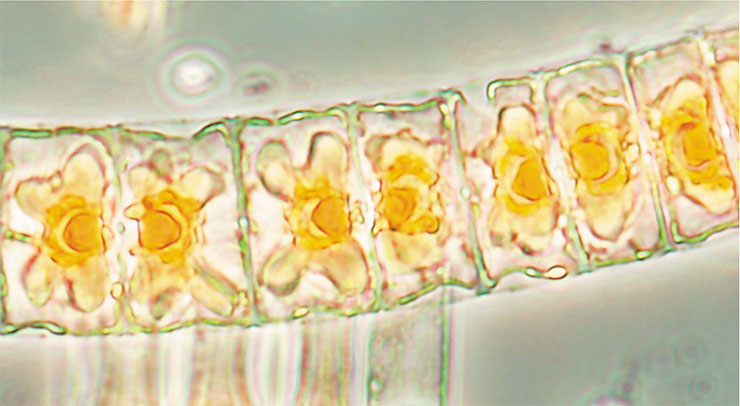
It is established that algae likewise “cry” with oxylipins in response to infection, stress, or injury and use them for immune defense. But while we have detailed biosynthesis charts for these bioactive compounds in plants and animals, our knowledge about algae is limited to the fact they have lipoxygenase(–s) with a particular substrate specificity (Andreou, Brodhun, Feussner, 2009).
For example, we may only guess which pathway is used by brown and diatom algae for the biosynthesis of pheromones, i. e., the signal compounds enabling male and female gametes to find each other, which represent a remarkable instance of chemical communication in unicellular organisms. In terms of chemical structure, these molecules are branched or cyclic hydrocarbons, and to date, we have strong evidence that they are formed from oxylipins (like volatile aldehydes and alcohols in plants) (Rui, Boland, 2010).
Speaking about such a kingdom of multicellular life as fungi, their oxylipin biosynthesis is very complicated and is not fully clear even to specialists.
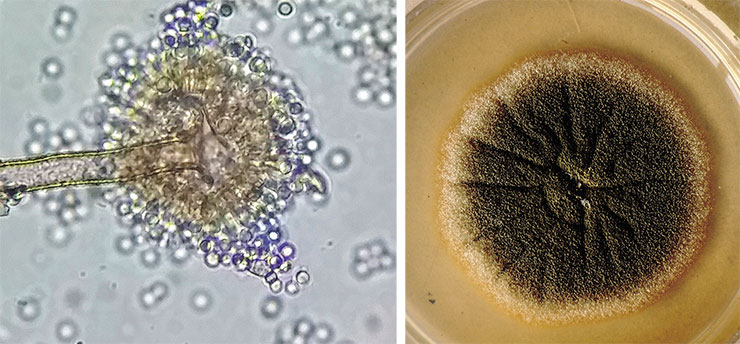
It is known that fungi do possess lipoxygenase enzymes. However, some of them contain manganese ion instead of the “classical” iron, but it has no significant influence on their biochemical mechanism of action (Brodhun, Feussner, 2011). Fungi, like plants, also synthesize volatile oxylipins.
However, these wonderful organisms, blending both animal and plant traits, also have homologues of our cyclooxygenases. But their function in fungi is by no means to synthesize prostaglandins. These enzymes either do the same work as lipoxygenases do (convert fatty acids into hydroperoxides) or fix two hydroxyl groups into a fatty acid molecule at once (diol synthases do this way).
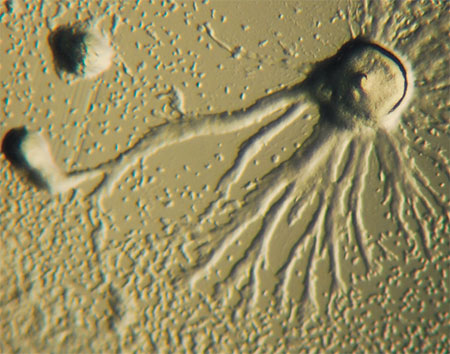 Fungal oxylipins perform an important function providing quorum sensing for the cells and thus enabling them to change their morphology depending on population density. Fungi do not “cry” with oxylipins – they signal “Don’t push!” or “Leave me alone!” with them. A neighbor can respond by switching from a vegetative form consisting of long filaments (hyphae) to a more compact or even unicellular form (as in the case of yeast). This is the way Ceratocystis ulmi, the fungus causing Dutch elm disease, acts (Brodhun, Feussner, 2011). Conversely, a mould fungus Aspergillus flavus can react to oxylipin action by changing its asexual reproduction strategy, switching from resting sclerotia to spore-forming conidia (Horowitz Brown et al., 2008).
Fungal oxylipins perform an important function providing quorum sensing for the cells and thus enabling them to change their morphology depending on population density. Fungi do not “cry” with oxylipins – they signal “Don’t push!” or “Leave me alone!” with them. A neighbor can respond by switching from a vegetative form consisting of long filaments (hyphae) to a more compact or even unicellular form (as in the case of yeast). This is the way Ceratocystis ulmi, the fungus causing Dutch elm disease, acts (Brodhun, Feussner, 2011). Conversely, a mould fungus Aspergillus flavus can react to oxylipin action by changing its asexual reproduction strategy, switching from resting sclerotia to spore-forming conidia (Horowitz Brown et al., 2008).
All these transformations are driven by lipoxygenase-derived oxylipins. Meanwhile, “distant relatives” of our cyclooxygenases participate in the synthesis of oxylipins that provide for the switching between sexual and asexual reproduction (Brodhun, Feussner, 2011).
Protozoa and bacteria: more questions than answers
The higher eukaryotes (organisms possessing a shaped cell nucleus) of all the aforementioned kingdoms use oxylipins for cell-to-cell-signalling. Regardless of what a cell “wants to say”, it sends some important oxylipin message to another cell. In other words, the principal and common function of oxylipins is to serve as a kind of a biochemical «Esperanto».
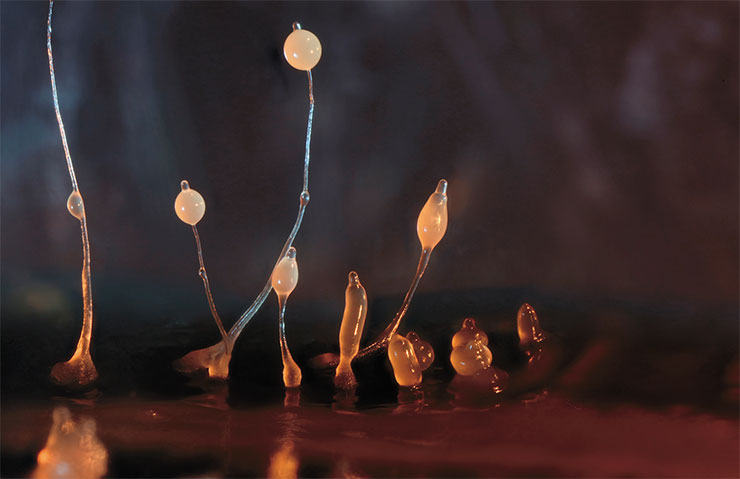
It could be an example of spectacular evolutionary conservation if we knew a little more about oxylipin biosynthesis enzymes beyond plants, animals, algae, and fungi – but we do not. Our knowledge gap spans the evolutionary root of these large groups. It is discouraging because, as Theodosius Dobzhansky (one of the founders of modern evolutionary biology) wrote, “nothing in biology makes sense except in the light of evolution.”
In this regard, we know very little both about bacteria (prokaryotic organisms) and about protozoa (unicellular or colonial eukaryotes incapable of photosynthesis).
The “slug’s” cells begin to differentiate and eventually form different parts of the fruiting body, which is very similar to a cap mushroom in its mature form. The life cycle of this pseudo-multicellular organism ends with the dispersal of spores, which germinate into new amoebae. Slime moulds whose lipoxygenase is “switched off” cannot form a fruiting body
There are no published data on protozoan lipoxygenases with the exception of a database record on a slime mould Dictyostelium discoideum (Phenotype and Strain Details for lipA-//dictyBase [Electronic resource]). Satisfied dictyostelium is a usual soil amoeba. But upon starvation, the amoebas crowd together to form a multicellular aggregate moving as an integral whole and later forming a fungus-like fruiting body. And, if a dictyostelium has a lipoxygenase gene mutation, fruiting body formation is disrupted.
Regarding bacteria, lipoxygenase biochemistry has been described for several species of cyanobacteria and myxobacteria. Like in the previous cases, these enzymes convert fatty acids to hydroperoxides, which are further transformed to hydroxides. But it is not yet possible to provide a detailed outline of oxylipin biosynthesis in bacteria, as well as in algae.
It is worth noting that lipoxygenases are conveniently conserved – in almost all living organisms, this family of enzymes performs the same functions. Moreover, all of these enzyme proteins are based on a conserved fragment (domain), which is easily recognizable in amino acid sequence analysis by special software. The occurrence of these enzymes can therefore be estimated simply by “scanning” existing databases with bioinformatics tools.
This analysis has detected genes encoding potential lipoxygenases in 0.5 % of the bacterial genomes “deciphered” to date. In archaea, the most ancient unicellular organisms without nuclei and membrane structures, which were recently separated from bacteria into a distinct superkingdom, this method has not revealed the presence of lipoxygenases (Horn et al., 2015; Kurakin, Samoukina, Potapova, 2020).
These results show that most bacteria live normally without lipoxygenase although hundreds of them do have this enzyme. And it is almost the same enzyme that produces leukotrienes in our bodies and jasmonates in a geranium plant on the window sill. But information about its function in bacteria is extremely scarce.
Back in 2007, German researchers tried to injure the cyanobacterium Nostoc punctiforme with ultrasound in the same way that other research groups had previously done with diatom algae. It turned out that in doing so, Nostoc, like diatoms, «cries» with oxylipins, the products of lipoxygenases (Lang, Feussner, 2007). Unfortunately, this study did not look into the response of other bacterial cells to the release of these molecules and their possible biological role.
In contrast, studies of the soil bacterium Myxococcus xanthus examined this one response only – adding the bacterium’s own oxylipin to the culture increased cell motility (An, Oh, 2018). But it remains unclear what this behavior means under real conditions.
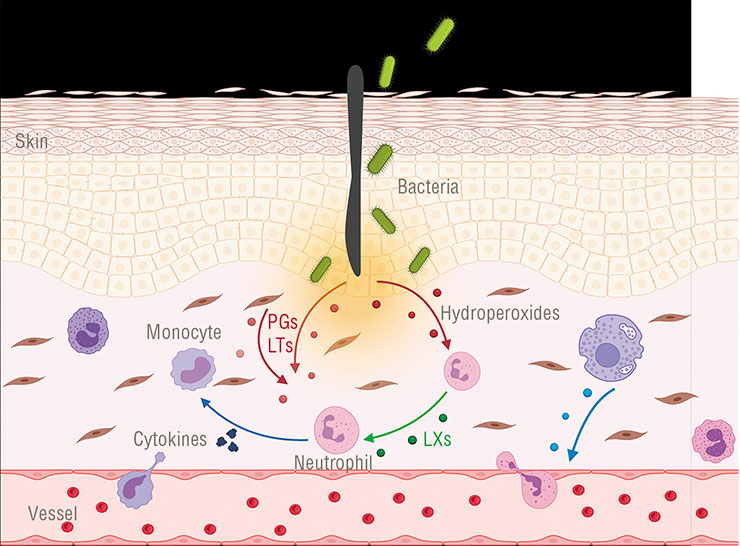
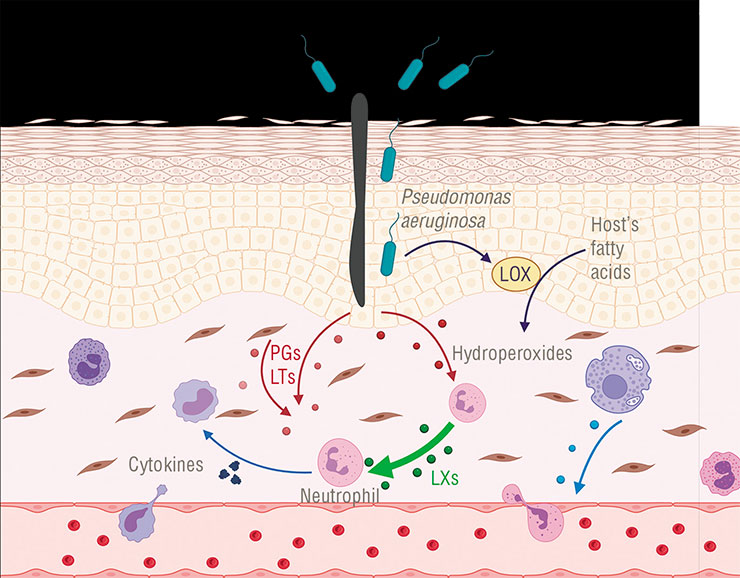
Lipoxygenase enzymes involved in oxylipin synthesis were found in some bacteria, but little is known about their function. An exception is Pseudomonas aeruginosa, which uses this enzyme to activate the synthesis of anti-inflammatory oxylipins in infected cells. It is possible that other pathogenic bacteria use this “trick” as wellAgainst this background, the information about the function of lipoxygenase and oxylipins in Pseudomonas aeruginosa, a dangerous nosocomial pathogen, is a breakthrough. This pathogen was found to use lipoxygenase and oxylipins to skillfully evade the infected human’s immune response (Morello et al., 2019).
How does it work? Let us recall that we have several groups of anti-inflammatory eicosanoids, including lipoxins, which serve to terminate the injury response in time. And Pseudomonas aeruginosa uses its lipoxygenase to steer the host’s eicosanoid synthesis in the intended direction! It does so by producing lipoxin precursor compounds from our lipids since it has no lipids of its own. These molecules are then captured by human macrophages (a kind of immune cells), in which further synthesis of lipoxins takes place. Lipoxin molecules are released through the cell membrane when “ready”. Other immune cells at the pathogen invasion site perceive them as an «Attack over!» signal and disable their defense activity, so that the bacterium could keep doing its nefarious deed undisturbed.
Still, Pseudomonas aeruginosa is only one of the many “lucky” bacteria that possess lipoxygenase. Now, let us return to the question: Why does the bacterial world need lipoxygenases, or rather the oxylipins that are synthesized with them?
Bioinformatics lifts the veil
 We applied bioinformatics methods to puzzle out why life needs oxylipins and how it learned to produce them. The main target was lipoxygenase, i. e., the most conserved oxylipins biosynthetic enzyme, as already mentioned above. We “scanned” the bacterial and protozoan protein sequences available in databases to find lipoxygenases among them. Then we started searching for connections between the presence of this enzyme and specific traits of its owner (Kurakin, Samoukina, Potapova, 2020).
We applied bioinformatics methods to puzzle out why life needs oxylipins and how it learned to produce them. The main target was lipoxygenase, i. e., the most conserved oxylipins biosynthetic enzyme, as already mentioned above. We “scanned” the bacterial and protozoan protein sequences available in databases to find lipoxygenases among them. Then we started searching for connections between the presence of this enzyme and specific traits of its owner (Kurakin, Samoukina, Potapova, 2020).
We started from a simple statistical analysis by counting lipoxygenase occurrences in different taxonomic groups. At this stage, we already identified the leading taxa both among bacteria and among protozoa. Among bacteria, myxobacteria (Myxococcales) as well as Nostocales and Oscillatoriales from the cyanobacterial phylum were particularly prominent. Among the protozoans, there were slime moulds (Mycetozoa) and water moulds (Oomycota). All these taxa share one important trait, i. e., multicellularity. There is no mistake – multicellular bacteria and protozoa (like the aforementioned Dictyostelium slime mold) do exist.
Let us start with cyanobacteria. Many of them look like a filament under the microscope and resemble filamentous green algae in terms of physiology. All the cells in the filament are connected into what looks like a single organism, with special bridge-like gap junctions forming between the cells for them to exchange metabolites. Moreover, the cells of Nostocales colonies can even differentiate into different types: vegetative cells, which form the filament itself, and heterocysts, capable of fixing nitrogen.
 Myxobacteria and slime moulds are counterparts, in some way, although they belong to different domains – they have similar types of multicellularity and ecological niche. These organisms live in forest cover, in soil, on decaying leaf litter and wood; they prey on other bacteria. When food is scarce, myxobacteria swarm together to prey collectively; when they begin starving to death, they gather into a single multicellular organism to form fruiting bodies. Unicellular slime moulds are able to do almost the same things.
Myxobacteria and slime moulds are counterparts, in some way, although they belong to different domains – they have similar types of multicellularity and ecological niche. These organisms live in forest cover, in soil, on decaying leaf litter and wood; they prey on other bacteria. When food is scarce, myxobacteria swarm together to prey collectively; when they begin starving to death, they gather into a single multicellular organism to form fruiting bodies. Unicellular slime moulds are able to do almost the same things.
On the other hand, oomycetes in their life form completely mimic… mushrooms. These pseudo-fungi even form a mycelium consisting of filaments, although in terms of biochemistry and phylogeny, they are closer to certain algae. Thus, the leaders in lipoxygenase occurrence are those taxa that evolution was “experimenting” upon to create multicellularity, albeit primitive one.
New evidence supporting this hypothesis came from phylogenetic analysis, the main idea of which is to reconstruct the evolution of proteins (or nucleic acids) on the basis of their sequences. Such phylogenetic models usually have a tree-like appearance, but we preferred to build them in the network-like way since we had reasons to assume that lipoxygenase genes spread among bacteria horizontally (between different species) rather than vertically (from a common ancestor to its descendants).

Our intuition did not deceive us. It turned out that bacteria transferred lipoxygenases to each other mainly by horizontal gene transfer. This became clear as soon as we revealed that evolutionarily distant groups fell into the same phylogenetic clusters. Given the relatively low prevalence of multicellularity in the bacterial world, it was arguably not a coincidence.
Additional analysis showed that lipoxygenases most likely first appeared in cyanobacteria. These organisms became the first beings to learn how to form multicellular structures. Most likely, they began doing that as early as the time of the so-called Great Oxidation Event, i. e., the appearance of free oxygen in the atmosphere about 2.5 billion years ago. The genes for lipoxygenases were repeatedly “borrowed” from cyanobacteria by myxobacteria, another group of multicellular bacteria.
In order to reconstruct lipoxygenase evolution in eukaryotes, we considered the statistical significance of each branch, the molecular evolution of the respective taxa, and other factors. The data were brought together to identify the putative events of lipoxygenase acquisition in different groups of multicellular eukaryotes on the phylogenetic tree. It turned out that there were several such events! All of them were mutually independent and, most importantly, evolutionarily related to the times of emergence of multicellularity.
Bacteria: genial villains
Additional statistical and evolutionary analysis also gave us some surprising results. It seems that not only Pseudomonas aeruginosa uses lipoxygenase to invade the human body. There may be many such bacteria, which greatly increases the scale of the problem.
Many species in our sample do not exhibit multicellularity in any form. When we grouped them out according to their ecological traits, we found among them many symbionts and human pathogens.
Alongside Pseudomonas aeruginosa, many of its counterparts have been found to belong to the “pathogenic” group. These bacteria pose little danger to healthy people but great danger to those with compromised immune systems or co-morbidities. Almost all members of this bacterial group are a common cause of nosocomial infections and produce similar lesions in patients: sepsis, surgical skin infection, subcutaneous tissue lesions, and lung diseases.
Another characteristic feature of bacteria in this group is a broad host range. Many of these bacteria were even previously described as cross-kingdom pathogens, which means their ability to infect hosts from different kingdoms. For example, Pseudomonas aeruginosa is capable of attacking plants as well as humans. It is not alone in this ability – e. g., the bacteria Burkholderia gladioli and Pantoea ananas can do this too. Their very names tell us exactly which plants these bacterial species, known to be dangerous nosocomial pathogens, prefer to infect.
According to our data, the lipoxygenases of all pathogens and symbionts cluster together when analyzed. Perhaps, this is not a coincidence. Most likely, these bacteria share with Pseudomonas aeruginosa its invasion strategy; i. e., they synthesize oxylipins to «deceive» the host’s immune system.
 This role of these signal molecules is also a form of communication – and another aspect of the biochemical “Esperanto”. Notably, this aspect might still be a secondary one in terms of evolution as bacteria first invented lipoxygenase to maintain cell-to-cell oxylipin signalling, and this “language” struck roots in a number of bacteria and in many eukaryotes. It was only later that a small group of bacteria learned to use it for deception and fraud, generously sharing it with other “villains”'.
This role of these signal molecules is also a form of communication – and another aspect of the biochemical “Esperanto”. Notably, this aspect might still be a secondary one in terms of evolution as bacteria first invented lipoxygenase to maintain cell-to-cell oxylipin signalling, and this “language” struck roots in a number of bacteria and in many eukaryotes. It was only later that a small group of bacteria learned to use it for deception and fraud, generously sharing it with other “villains”'.
The following picture emerges from our bioinformatic study. Lipoxygenases, the key enzymes in the synthesis of signalling oxylipin molecules, most likely were “invented” by cyanobacteria. Other multicellular bacteria – myxobacteria – borrowed them. Some groups of eukaryotes also adopted lipoxygenases (from whom exactly, is still unknown) during those periods of their history when they acquired multicellularity, albeit primitive one. This pattern can be traced throughout the living world. It explains why only 0.5 % of bacteria have lipoxygenases while their occurrence is much higher in multicellular plants and animals.
Here another question arises: If lipoxygenases accompanied the acquisition of multicellularity, what role did they play in this process? There is no definite answer to this question yet. The established connection only supports the hypothesis that bacteria use lipoxygenase to synthesize oxylipins in the same way as plants and animals do and that they were the authors of this idea. But how critical was such communication for the emergence of multicellularity? This question is quite intriguing: What if this is where the key lies to unlocking the complexity of modern living world?
Studying oxylipin signalling evolution and lipoxygenases themselves also opens up new possibilities of using these enzymes as genetic markers of dangerous bacterial strains or as a target for overcoming antimicrobial resistance. And we are not alone in these conclusions.
Oxylipin signalling used by humans and animals for cell-to-cell communication was studied in bacteria. Hopefully, this study will help develop strategies to combat antimicrobial resistanceIn 2019, a team led by J. Campos-Gomez from the Southern Research company (United States) experimentally studied another oxylipin communication system in Pseudomonas aeruginosa, which involves a diol synthase enzyme, not related to lipoxygenase (Martínez, Cosnahan, Wu, et al., 2019). The fact that bacterial pathogens need quorum sensing to successfully invade human organism was known before, but it was usually associated with other chemical compounds. Now we know that oxylipins are also involved in this form of bacterial communication, and targeting them may be the principle of action of new antibiotics.
Later, a piece with an encouraging title “New bacterial signaling language offers pathway to treat infections” appeared in Infection Control Today. There, Campos-Gomez stated: “We are trying to develop a new generation of antibiotics that do not directly kill the bacteria, reducing the odds that it will develop resistance to the drug. We want to disarm the bacteria, so that the immune system takes care of the bacteria itself.” And this statement surprisingly echoes our piece in Laboratory News, titled “Microbial lipoxygenases: the next target to fight antibiotic resistance?” (Kurakin, Samoukina, Potapova, 2020).
We hope that the results of our research on oxylipin signalling evolution, during which we made such a coincidental finding as an association with bacterial pathogenicity, will attract the attention of our colleagues in science and medicine to lipoxygenases, and our work will become an example of basic research that unexpectedly yielded a practical output.
References
Andreou A., Brodhun F., Feussner I. Biosynthesis of oxylipins in non-mammals // Prog. Lipid Res. 2009. V. 48. N. 3/4. P. 148–170.
Brodhun F., Feussner I. Oxylipins in fungi // FEBS J. 2011. V. 278. N. 7. P. 1047–1063.
Freeman B. C., Beattie G. A. An overview of plant defenses against pathogens and herbivores // Plant Health Instructor. 2008.
Heldt H.-W., Piechulla B. Plant Biochemistry. Elsevier, 2011. 622 p.
Horn T., Adel S., Schumann R. et al. Evolutionary aspects of lipoxygenases and genetic diversity of human leukotriene signaling // Prog. Lipid Res. 2015. V. 57. P. 13–39.
Kurakin G. F, Samoukina A. M, Potapova N. A. Bacterial and Protozoan Lipoxygenases Could be Involved in Cell-to-Cell Signaling and Immune Response Suppression //Biochemistry (Moscow). 2020. V. 85. N. 9. P. 1048–1063.
Morello E., Perez-Berezo T., Boisseau C. et al. Pseudomonas aeruginosa lipoxygenase LoxA contributes to lung infection by altering the host immune lipid signaling //Frontiers Microbiol. 2019. V. 10. P. 1826.
Savchenko T. V., Zastrijnaja O. M., Klimov V. V. Oxylipins and plant abiotic stress resistance // Biochemistry (Moscow). 2014. V. 79. N. 4. P. 362–375.
https://www.infectioncontroltoday.com/view/new-bacterial-signaling-languageoffers-pathway-treat-infections
https://www.labnews.co.uk/article/2031213/microbial-lipoxygenases-a-nexttarget-against-antibiotic-resistance
Based on an article in the ‘Biomolekula’ (Biomolecule) online publication as part of the ‘BioMolText 2020/2021’ competition