The Genome’s Dark Matter
An avalanche development of DNA sequencing methods during the last decade has led to an explosive increase in the number of decoded genomes. It has become evident that for many years researchers underestimated the role of noncoding genomic regions, i. e., the genomic fragments lacking any genes yet actively involved in controlling the living cell metabolism. “Science” magazine considers the discovery of this phenomenon, referred to as the genome’s “dark matter”, one of the outstanding advances of the last decade
Long ago such terms as “DNA,” “gene,” “genome,” and the like moved from research papers to newscasts, which is quite natural. How can we feed many billions of the world population? How can we cure cancer and AIDS? How can we extend the human life? These are but a few ambitious challenges to modern biology. Presumably, a similar situation took place in the middle of the last century, when advances in nuclear physics were believed to help create boundless sources of free energy. Unfortunately, many of these problems yet remain unsolved, and the society is running out of patience.
The concept referred to as the central dogma of molecular biology was formulated by Francis Crick as early as 1958. Elegant in its simplicity and self-consistency, it explains how hereditary information is preserved in a line of generations, how it is arranged in genes, and how it is transformed into protein molecules via RNA, eventually taking the shape of an individual living being. At that time, they believed that availability of a rapid and inexpensive “reading” (sequencing) of DNA sequences would make it feasible to describe the overall genome by merely “reading” one gene after another.
However, the first “draft” version of the human genome, published in 2001, demonstrated that it contained far fewer genes than it was anticipated (21,000 instead of 100,000). In other words, genes per se cover only 1.5 % of the total length of genomic DNA. The remaining part of the genome was regarded as dead weight. The apparent simplicity of genome organization gave hope for a quick breakthrough in biology, although it had already been found that the regulatory mechanisms involved in gene expression (activation of gene operation) were considerably more intricate than it was earlier expected.
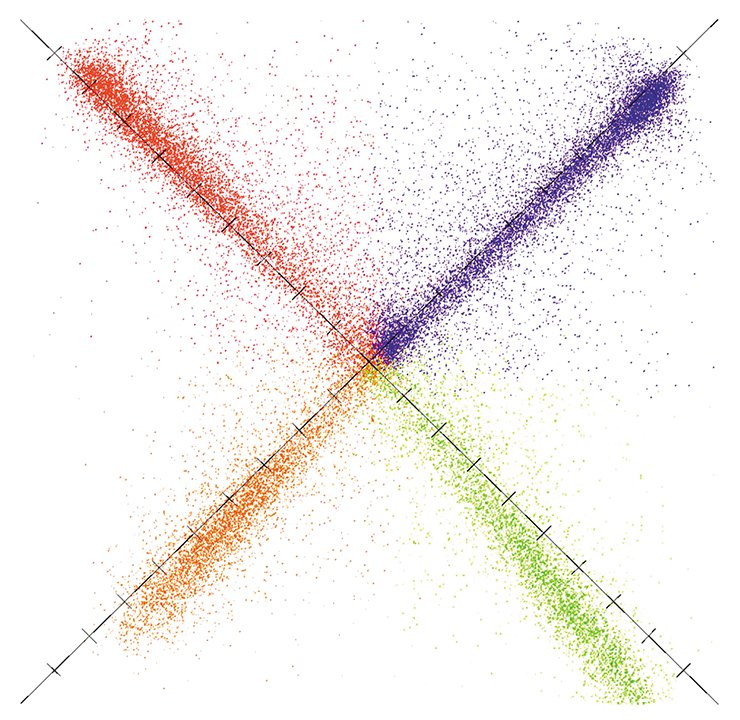
Finally, the sequencing of tens and, later, hundreds of various genomes in the early 21st century made it clear that the tremendous arrays of dead-weight DNA were literally saturated with various regulatory sites. Moreover, it was discovered that chemical modifications of nucleotides, the “letters” of genetic code, play an exeptionally significant role in the spatial arrangement of a DNA molecule in the cell nucleus, in the regulation of gene expression, and so on.
It was this particular complexity of the mechanisms regulating the processes described long ago as the central dogma of biology that formed the background for the concept of the genome’s dark matter, i. e., the set of all its noncoding elements, earlier regarded as insignificant auxiliary elements. Although the epithet “genome’s dark matter”, used in a Science publication (Pennisi, 2010), sounds rather alarming, all the effects mentioned above have been known for a long time; what appeared unexpected was the scale of their significance. Another reason explaining the phenomenon of genome’s dark matter is that in the technologies for experimental data processing and visualization are lagging behind the technologies for obtaining these data: in the former activities, we frequently continue to use the ideology and approaches of the “pregenomic” era.
The most interesting discoveries of the postgenomic era are associated with the roles and functions of the DNAs’ “sister”, RNA. Earlier, these molecules were assigned a minor role of information “transmitters” from DNA to proteins. The situation changed in the 1990s with the discovery of RNA interference phenomenon, that is, the inhibition of gene expression at the stage of information transcription or protein synthesis. Currently, it is known that about 80 % of all DNA in the cell are “rewritten” in RNAs; moreover, the overall diversity of functions fulfilled by various RNA classes is yet to be described.
For many years the Institute of Chemical Biology and Fundamental Medicine, Siberian Branch, Russian Academy of Sciences (Novosibirsk, Russia) has been involved in studing the role of RNA in manifold physiological processes in normal and pathological cases, including some types of cancer. Massively parallel sequencing technologies make it possible not only to attain any level of detail, required, for instance, for identification of rare RNA species, but also to free such studies from our initial views about a particular phenomenon.
For example, a set of RNA fragments with an unexpectedly high stability has been discovered in blood plasma, whereas most of the RNA molecules degraded rapidly. Interestingly, a set of such unusual RNAs depends on sex and age; moreover, it may change if there are pathological processes. These RNAs, “messengers” of the genome’s dark matter, are promising tools for an early diagnosing of human and animal diseases.
References
Pennisi E. Shining a light on the genome’s ‘dark matter’ // Science. 2010. Vol. 330(6011). P. 1614.