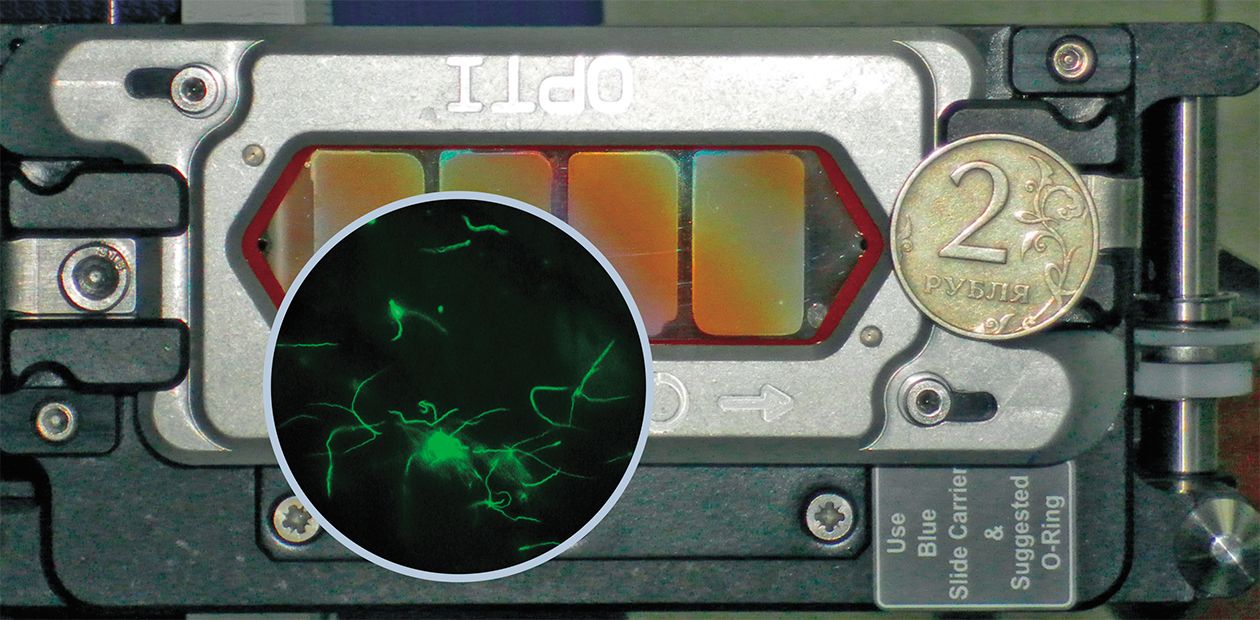
Sequencing the Genome of Borrelia, Agent of Lyme Disease
In 2011, the complete genome of the pathogenic Borrelia species, typical for Russia, was isolated from a taiga tick at the Tomsk scientific and production association NPO Virion and then sequenced at the Institute of Chemical Biology and Fundamental Medicine, Siberian Branch, Russian Academy of Sciences (Novosibirsk, Russia)
The studies of the tick-borne diseases are becoming ever more topical: in 2011 alone, about half a million tick bites were recorded in Russia. Via tick bites, humans can get not only the widely known viral tick-borne encephalitis, but also another, no less dangerous malady, tick-borne borreliosis, or Lyme disease.
Borreliosis is a polysystemic disease potentially affecting the skin, locomotor apparatus, as well as the nervous and cardiovascular systems. If the necessary treatment is not started immediately, the disease turns into a chronic form, which requires longer and more complex treatment, not always effective: the patient runs the risk of developing a disability because of severe injuries of tissues and organs. Unfortunately, no vaccines preventing this disease are currently available.
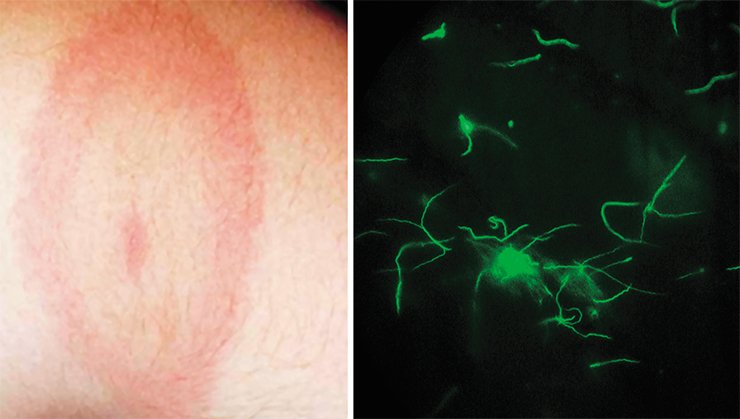
In the case of borreliosis timely medical assistance means an immediate and accurate molecular genetic diagnosing of the disease. However, this is not a simple task since in different regions there are different Borrelia species pathogenic for humans; moreover, these species may display a high intraspecific polymorphism. Thus, this interferes with using in Siberia the diagnostic kits developed in the United States or in Europe.
The key point in designing state-of-the-art diagnostic, preventive, and therapeutic tools to treat the disease is studying the genome (a complete set of hereditary material) of its causative agent. An ideal method is sequencing (determining the sequence of nucleotides in DNA), the complete genome of the infectious agent. Correspondingly, elaboration of diagnostic kits, vaccines, and therapeutic tools specific and efficient primarily for the Borrelia species typical of Russia requires that the genomes of local genetic variants of these bacteria be sequenced. However, until recently such projects were very expensive and labor-consuming.
Development of sequencing methods, begun as early as the 1960s—1970s, had as its final goal a routine decoding of complete genomes without spending too much time or money. The first considerable advance in this field was the Sanger method of enzymatic sequencing, which allows a reliable determining of sequences up to a thousand nucleotides in one experiment. Having undergone numerous modifications, this method is still a “draft horse” of sequencing.
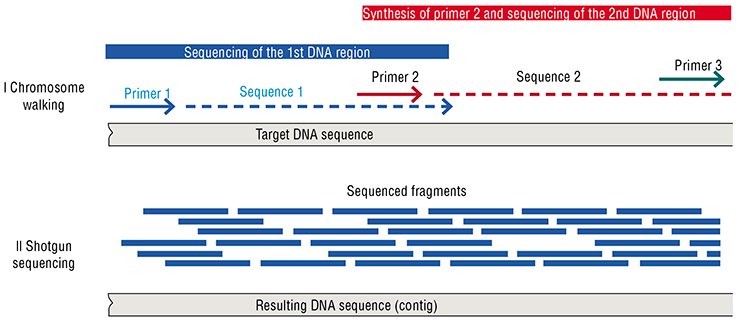
The essence of the second sequencing strategy, referred to as a shotgun method, is that the target DNA is initially fragmented in a random fashion. The fragments are cloned into vector DNA molecules, multiplied, and sequenced with the use of a universal primer. The target DNA sequence is then deduced from the overlapping sequences of short fragments. In this approach, the total length of the determined sequences usually exceeds that of the target DNA several times
In order to simplify and speed up the sequencing of extended DNA regions, a strategy referred to as “shotgun sequencing” was proposed. Its essence is that the initial DNA sequence (contig) is determined by decoding multiple overlapping short fragments. However, this strategy implies that the total amount of the data obtained should several times exceed the length of initial sequence. Apparent simplicity and elegance of this approach concealed an all-out commitment of the researchers, for it was proposed and tested at the time when the determining of one nucleotide cost several dollars. In addition, neither effective algorithm for assembling the target DNA from short sequences nor powerful enough computers able carry out the assembly were yet available. No wonder that the sequencing of the human genome begun in the 1990s took 13 years of coordinated efforts of hundreds of scientists from dozens of research institutions of various countries.
The situation changed drastically in the 21st century, when fundamentally new massively parallel sequencing technologies were developed based on the accumulated experience. Such currently existing technologies utilize different basic processes; however each of them has the same feature: sequencing is concurrently conducted in millions of microscopic reaction sites. Thus, millions of individual short sequences are determined in the same experiment and then used to assemble extended DNA sequences by the shotgun method. Note that time, reagent consumption and, eventually, money spent per one nucleotide are by several orders of magnitude less as compared with traditional Sanger sequencing. Thus a rapid and inexpensive sequencing of complete genomes has become a reality.
Researchers at the Novosibirsk Institute of Chemical Biology and Fundamental Medicine have considerable experience in this field: in 1989 they sequenced the genome of the tick-borne encephalitis virus. The genome of Borrelia, though, is more than a hundredfold longer and has a far more intricate structure.
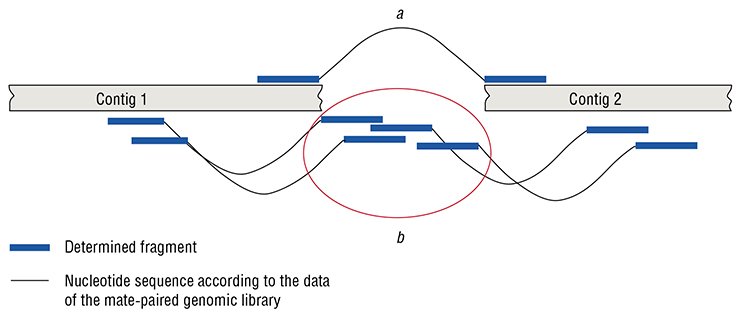
Genomes of even the simplest organisms, such as Borrelia, usually contain several replicons (hereditary units autonomously replicating themselves during cell division) – either chromosomes or plasmids. As a rule, a genome also contains regions copied several times (satellite DNA, ribosome genes, and others). As a result, the target nucleotide sequences, obtained by the shotgun method are shorter than a complete sequence should be.
The sequences “interrupt” just at the sites containing lications. In several cases, decoding such genomes is theoretically possible but requires traditional Sanger sequencing of millions of sequences, i.e. tremendous additional resources. That is why a genome is often declared finished long before the continuous reliable sequences of all its replicons are determined. For example, the human genome is still represented by hundreds of nucleotide sequences covering approximately 95 % of its real length.
However, this situation can sometimes be resolved quite easily with the help of the so-called “mate-paired genomic libraries”. Each molecule in such a library has two different “tags” of the target genome at its ends. The distance between these tags is known and specified by the researcher. Sequencing such libraries makes it possible to arrange the preassembled contigs in the correct order and orientation, and in some cases to fill the gaps between them.

To improve the sequencing quality, it is optimal to use a combination of various methods, each of which possesses unique advantages. The Siberian Borrelia species (B. garinii BgVir), isolated at the scientific and production association NPO Virion, was determined at the Institute of Chemical Biology and Fundamental Medicine by the massively parallel sequencing method that used a Roche platform (a genomic fragment library) and a SOLiD platform (a mate-paired genomic library).
The Roche pyrosequencer can determine long (about 400 bp) nucleotide sequences and makes it possible to sequence the structures of the genomic regions that are absent in the already decoded genomes of even the most closely related species. As for the SOLiD platform, it provides for an arbitrary large coverage of the genomes of any size at a minimal cost, and in combination with mate-paired libraries, it sequences the regions not covered with the data for other platforms.
This approach allowed the Novosibirsk researchers to use the advantages and minimize the shortcomings of each platform. The final sequence assembly, search for genes, and analysis of the differences between the sequenced genome and the known genomes were performed on the basis of the facilities of the Joint Access Center Bioinformatics, using the software algorithms developed at the Institute. Comparative analysis involving other Borrelia species has uncovered several structural distinctions including the genes coding for surface proteins and some enzymes; it has also determined a set of genes unique for the genome of B. garinii BgVir.
Experience accumulated by the Siberian researchers in the sequencing of simple viral and bacterial genomes allows them in future not only to decode viral and bacterial genomes in the future, but also to tackle more intricately arranged genomes of higher organisms.
E. V. Brenner, Candidate of Biology;
A. M. Kuril’shchikov; and N. V. Fomenko, Candidate of Biology
(Institute of Chemical Biology and Fundamental Medicine, SB RAS, Novosibirsk)
References
Fomenko N. A. Kleshhevoj borrelioz: bolezn’ na vsju zhizn’? // Nauka iz pervyh ruk. 2007. № 3 (15). S. 44—51.
Fomenko N. A. Tick-borne borreliosis: a life-long disease? // Science first hand. 2007. N 3 (15). P. 44—52.