To be born a Baobab
Envy is perhaps the most powerful factor of biological and social evolution of the man. When a man sits down in the shade of a baobab and wonders: “When our tribe first came here, this old tree was already here. My great-grandfather used to sit here, so did my grandfather and father. Now I’m here. My son will sit here, and so will my grandson, and so on. When everyone dies, the tree will still be here. This is so upsetting…” Such envious interest of humankind to long-living species is understandable: what if we discover something that will let us live longer – like the baobab? This is why scientists have such a keen interest in all of nature’s models of slowed as well as accelerated aging. Like with many other natural phenomena, the more distant the relation of these models stand to conventional laboratory animals, the more amazing the mechanisms they use in dealing with aging
Aging as a term is most often applied to multicellular organisms. Most single cells organisms are considered potentially immortal: during division, they split equally into two organisms identical to the parent. However, in recent years even some bacteria were found to undergo unequal division processes. For instance, Caulobacter crescentus, a species of bacteria inhabiting freshwater habitats, divides asymmetrically, forming a large cell attached to the substrate and a smaller cell that moves freely in the water. Eventually, the division rate of the first “maternal” cell slows down, thus showing signs of aging (Ackermann et al., 2003).
When baker’s yeast divide, small cells bud from a large maternal cell; and, after a number of divisions, the maternal cell dies. Signs of aging in the yeast cells include increase in size, loss of turgor (intracellular osmotic pressure), lengthening of the cell cycle, reduction of protein synthesis, accumulation of extrachromosomal DNA, etc. (Clay and Barral, 2013). Interestingly, maternal cells selectively pass their most efficient mitochondria on to their daughters (mitochondria are organelles responsible for energy production in the cell).
In another species of yeasts, called brewer’s yeast, cells divide symmetrically and are potentially immortal, – but only when the conditions are favorable. Thermal or oxidative stress causes all large and nonfunctional protein aggregates to accumulate in one of the daughter cells – and that cell eventually dies (Coelho et al., 2013). Therefore, even single cell organisms can serve as important natural models of aging research.
Multicellular organisms also have potentially immortal cells – the gametes; somatic cells, however, are programmed to die. One of the important features of aging in these organisms is the gradual loss of stem cells, which are capable of self-rejuvenation and can turn into all other types of somatic cells in the process of differentiation. Therefore, the loss of stem cells hinders the renewal of various tissues to the extent of making it completely impossible.
How the cell ages
Currently, researchers are putting their best efforts into understanding the aging process on the level of an individual cell. Let’s have a look at several crucial mechanisms involved in this process.
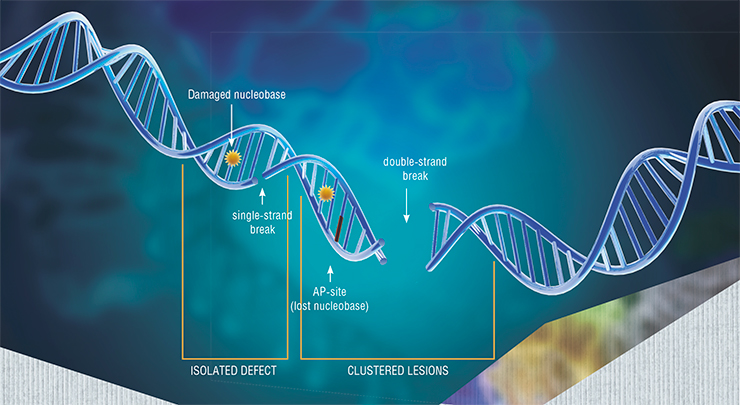
First, it is the corruption of the primary structure and epigenetic (i. e. not influencing the primary structure) status of the genomic DNA. A great number of factors, both external and internal, affect the genome, causing numerous mutations of the DNA: single nucleotide substitutions, double-strand breaks, chromosome rearrangements (e. g. inversions, when a fragment of the chromosome is reversed), shortening of terminal regions of chromosomes (telomeres), etc.
DNA repair systems successively cope with many of these defects – and how reliably they work defines the ability of the organism to resist natural damage to the genome. Weakening of the repair systems inalterably shortens life expectancy. Mitochondrial DNA is especially sensitive to mutagenic processes, since DNA repair in these organelles is weakened; moreover, they typically have higher concentrations of active forms of oxygen and other dangerous metabolites.
In recent years, epigenetic processes such as histone modification and DNA methylation were pointed out as important agents of cell aging (Han and Brunet, 2012). Histones are a family of nuclear proteins, which, among other things, participate in the packing of DNA strands. DNA methylation is the addition of the methyl group (–CH3) without changing the nucleotide sequence.
The second important mechanism of aging is the accumulation of proteins with incorrect structure. Aggregation of incorrectly folded proteins forming amyloids is associated with the development of age-related progressive neurodegenerative disorders, such as Alzheimer’s and Parkinson’s diseases.
Third, cell aging changes the so-called signal pathways – molecular mechanisms providing the transmission of signals within the cell, such as the so called insulin/insulin-like growth factor 1 signaling cascade, which affects a great number of cellular processes, such as energy exchange, stress response, etc. This pathway is involved in cell aging in a broad range of organisms, including coelenterates, roundworms, insects and mammals.
As for oxidative stress, which a number of research works have shown to contribute significantly to aging, the situation is ambiguous. Some experimental data show that small “portions” of oxidative stress produced during physical exercise or impulse fasting, on the contrary, seem to exercise antioxidative protection mechanisms – and, consequently, increase life expectancy.
Multicellular immortals
All multicellular organisms belong to the so-called eukaryotes – organisms with well-formed cell nucleus. Even among these higher organisms, one comes across the phenomenon of full potential immortality, typical for prokaryotes (bacteria and archaea).
Such “Koshcheis” (in Russian folklore, Koshchei is a wicked skeletal old man holding the secret of immortality) are found in some invertebrates that lack bilateral symmetry, such as hydroid polyps/jellyfish from the genus Turritopsis. Representatives of Turritopsis dohrnii are considered immortal as they do not show any signs of aging. In the “juvenile” phase, they are sedentary polyps, and during the next phase, when sexual reproduction occurs, they turn into a freely floating jellyfish. Amazingly, after reproduction, these organisms do not die, but instead, turn back into immature polyps. This cycle can repeat many times.

These multicellular animals always carry stem cells, which can differentiate into cells of different tissues and participate in the regeneration of any organ. This is what probably gives them their immortality. However, more complex life forms lost this ability in the course of evolution, most likely due to the necessity to prevent malignant transformation of cells into cancer cells.
At the same time, there is no apparent connection between the complexity of the organism and its lifespan. Sponges, which are among the most primitive multicellular animals, live up to 15 thousand years, while whales, who are mammals, only reach 200 years, and the rather simply organized roundworm, Caenorhabditis elegans, has no stem cells at all and only lives for a couple of weeks.

Multicellular organisms in general show a great diversity of forms, including some very whimsical creatures. For instance, arthropods, the most numerous group of animals, with all their metamorphoses, larvae, pupae, and imago, have all sorts of things going regarding the lifespan of each of these stages. Or plants with their peculiar biology. Trees, some of which can live for hundreds or thousands of years, like the one provided in the title of our article, which is a slight re-wording of a famous song by Vladimir Vysotsky.
But we will not be talking here about them. Next, we will take a look at some examples of the various types of longevity in vertebrates, to which humans belong, too. Many of these species can – or already have become living “models” for detailed study of the phenomenon of aging, useful when applied to our own longevity.
The short- and the long-living
Among vertebrates, fish are known for their longevity. The champion’s title here is held by the Greenland polar shark, a superpredator that can live up to 400 years. Representatives of this species reach sexual maturity by the age of 150. Polar rockfish, or sea perch (Sebastes) also live to 150 years and longer. Interestingly, these fish are viviparous. Other shark species are also long-lived, as well as sturgeons and catfish, reaching 50 to 150 years. At the same time, there are species of fishes which normally live for only a few months, such as species of the families Nothobranchiidae and Gobiidae (gobies).

Several years ago, researchers discovered that elephants carry several dozen copies of a gene coding a protein called p53 (humans have only two copies). This protein prevents cells with “unrepaired” DNA damage from dividing and launches the process of cell suicide, called apoptosis. It turned out that indeed, in elephants, apoptosis in response to radiation-induced DNA damage is more intensive than in humans.
Recent genomic analysis of 53 mammal species has highlighted another gene – LIF, a coding protein factor that inhibits the development of leukemia (Abegglen, Caulin, Chan et al., 2015). It turned out that while the majority of the studied organism only has a single pair of these genes, in the genomes of the African elephant, the manatee and the daman, there can be up to ten additional copies.
Accumulation of copies of genes is a widespread phenomenon. Such copies often carry errors and are non-functional; for this reason, they are called pseudogenes, or even “dead genes”. However, in elephants, a pseudogene called LIF6, somehow “came to life” – hence the nickname “zombie gene”.
Normally, LIF6 is virtually inactive, however, the p53 protein activates it when it “decides” to kill a cell. The protein coded by LIF6 disrupts the normal function of mitochondria, which are the “powerplants” of the cell, thus launching apoptosis, however, the mechanism is incompletely understood. Therefore, LIF6 together with p53 destroy potentially cancerous cells long before they begin to pose any real danger.
It looks like in the ancestors of the modern elephants, the LIF6 zombie gene came back to life about 25 to 30 million years ago, back when these animals were not that large. It is possible that it was the formation of this new, additional defense against cancer became the key element that allowed elephants to become the biggest and most long-lived mammals on land
Among the amphibians, the longest-living creature is the olm, or proteus – a blind salamander inhabiting some cold lakes and subterranean rivers in the Balkans. Among reptiles, it’s turtles – the Galapagos turtle can reach the age of 170, crocodiles, who can live over half a century, and Komodo dragons.
Many birds are relatively long-lived, too. The record holders are the Australian inca cockatoo, that lives over 80 years, and the Andean condor (79 years).

Among the unique order of the Monotremes, the echidna is noteworthy, as it can live up to half a century – twice longer than the platypus. Among the marsupials, the record is held by the wombat and the giant kangaroo, who can live up to 25–30 years.
Among the Afrotheria, a clade of placental mammals that formed independently in Afro-Arabia, the longest-living species are those who have adapted to aquatic lifestyle (manatees and dugongs) as well as elephants, who live up to 65 years (the number of natural predators of these species, due to their body size, is also limited). Elephants and manatees also have a remarkable ability to grow new teeth indefinitely, meaning a very highly retained potency of the corresponding tissues.
The afrotherian clade also includes tenrecs – small, hedgehog-like animals, the majority of which have been living in isolation on Madagascar since the Cretaceous period. It would be extremely interesting to compare the genomes, peculiarities of cellular differentiation and other characteristics of these animals, since their lifespan differs six-fold.

Among the Pilosa, sloths are relatively long-lived, reaching over 40 years. They spend most of their lives up in trees, rarely moving; when necessary, they crawl at a speed of 1 to 3 meters per minute. Their brain lacks convolutions and their digestive tract and bladder are constructed in such a way that they only need to empty them once a week. Even ocelots and harpy eagles, who are capable of hunting sloths in trees, rarely attack them. Truly a feast of a life, like a Diogene’s dream! At the same time, the lifespan of more active Pilosa, such as anteaters and armadillos, is almost twice as short as that of the sloths.
From a relatively recent discovery, we know that whales are in fact ungulates, who abandoned the dangers of terrestrial life for a more comfortable, and hence longer, marine existence. It is among the cetaceans there are long-living species such as the Greenland whale – the record holder of longevity among mammals (Moskalev et al., 2017).
WANT TO LIVE LONG? REPAIR YOUR DNA The first fact is pessimistic – in the cells of our body, DNA, the genetic material that controls the most complex vital processes, is damaged all the time. The second fact is optimistic – cells can protect their sacrosanct property with special DNA reparation systems. However, their efficiency decreases with age, and genomic modifications and other pathological changes accumulate within cells.Scientists have long suspected DNA reparation must play an important role in processes that grant longevity. It is known that mutations in genes coding reparation enzymes cause accelerated aging both in laboratory animals and in humans. At the same time, long-living animals have been found to be more resistant to genotoxic stress. However, we need direct proof of the link between the efficacy of DNA reparations and lifespan, and figure out the molecular mechanisms mediating this link.
This is not an easy venture, as there are many proteins involved in the reparation systems: both those that directly perform the repairs and those that take part in the process indirectly. The majority of attempts to improve the efficiency of reparation by increasing the activity of the corresponding enzymes have failed. The only exception was the SIRT6 gene, coding the sirtuin‑6 protein: increase in the activity of this gene improved the efficiency of reparation of double-strand breaks in DNA without negative side effects. The SIRT6 gene is often called the longevity gene, because laboratory mice with a knocked out SIRT6 live shorter lives than normal mice, and mice with a hyperactive SIRT6 live longer.
Recently, scientists evaluated reparation systems efficiency in 18 rodent species (Tian, Firsanov, Zhang, 2019). This mammal group is a convenient model for comparative studies of aging, because, despite their evolutionary proximity, they have extremely variable lifespan, ranging from 3 (mice) to 32 (beavers and naked mole-rats) years. Researchers examined not only the reparation of double-strand breaks in DNA, but also the excision reparation of nucleotides, which is used to remove extensive damage that distorts the double helix structure of DNA.
It turned out that the efficiency of excision reparation bears no obvious connection with lifespan. These data are not final, as the researchers compared species with different lifestyles. The main reason of extensive DNA damage is the ultraviolet radiation in sunlight, so nocturnal or diurnal character of a species’ activity could affect the selection vector.
The dire consequences of double-strand breaks may include complete termination of cell division or even cell death. If the breaks are repaired incorrectly, it may cause rearrangements not only in the sequences of specific genes, but in the whole structure of the chromatin, the foundation of the chromosomes. This can cause global disruptions in the function of genes and tissues – and organ dysfunction incompatible with long life.
Scientists discovered that the sirtuin‑6 protein, which participates in double-strand break reparation, is indeed more efficient in long-living rodents. Having analyzed its structure in the mouse and in the beaver, they found that the difference is limited to just five amino acids!
Using gene engineering, the researchers inserted the corresponding genes of the beaver or the mouse into human cells where the SIRT6 gene was “switched off” and obtained the expected results: cells with the “beaver” gene countered DNA damage more efficiently than cells that contained the “mouse” SIRT6 gene. In experiments with fruit flies, the favorite research object of geneticists, the scientists proved that not only was the “beaver” more efficient in countering DNA damage in a cell culture, but it also increased lifespan of the whole organism. The researchers are planning to study SIRT6 in species that live longer than humans, such as the Greenland whale
Hippos, who are the closest (more or less) terrestrial relatives of whales, are also long-lived. One should note that they are the most dangerous African mammals in terms of harm to humans. It is hippo encounters that claim the most human lives, much more than claws and fangs of any other African species. A while ago hippos were found to feed on fresh (and rotten) crocodile, antelope and lion meat – proceeding to graze peacefully along riverbanks and in marshes. They graze with their lips and only use their fearsome teeth for bloody fights and for attacks on other animals.
Odd-toed ungulates – horses, rhinos and tapirs – live long, too; among them, the horse lives the longest – up to 60 years.

Pinnipeds, who also fled to the sea from the dangers of land, are the longest-lived predators. Among pinnipeds, the Baikal seal lives the longest, as it has managed to get far away both from terrestrial and marine predators. Bears live long lives, too, and is there even anyone to cross the polar bear on its quest to live to forty-something? Felines, in general, live longer than canines.
Bats are a group that has radically cut all ties to their previous habitat and said “goodbye” to their terrestrial predators. The record here belongs to the Siberian Brandt’s bat, a tiny flying creature weighing no more than ten grams, which can live up to 40 years. The majority of other bats live much longer than their insectivorous ancestors and relatives.
As for the true Eulipotyphla (formerly Insectivora) – they are the ones who live fast and die young. Their lives are filled with round-the-clock sex and feeding frenzy – and are very short. Two venomous species, the Cuban and the Haitian solenodons, had life expectancies three times that of the shrews – until sailors brought dogs, cats and mongoose to their islands, who decimated their populations. They are almost on a par with hedgehogs, who carry a natural and efficient defense against predator attacks.
Among the lagomorphs, hares live almost twice as long as pikas and rabbits – and the mountain hare in particular (up to 18 years!).
THE SECRET TO BAT LONGEVITY? Among the bats studied up to date, mouse-eared bats (Myotis) are especially long-lived. These small animals, weighing anywhere from 2 to 45 grams, live over 20 to 30 years.A large research team has been studying four long-lived bat species for over 60 years by sampling tissues of several hundred marked individuals (Foley, Hughes, Huang et al., 2019). The researchers were especially interested in the length of the bat telomeres – the protective structures on the tips of chromosomes. In human cells, as well as in cells of most other mammals, telomeres shrink with each cell division; when they become too short, the cell ages and dies. Stem and cancer cells are exceptions – inside them, an enzyme called telomerase constantly builds up the shortening telomere regions of DNA. In normal cells, telomerase is inactive.
It turned out that in two species of mouse-eared bats – the Bechstein’s bat and the greater mouse-eared bat, telomeres do not shrink with age, and telomerase does not participate in sustaining their length. The shortening of telomeres is thought of as a defense mechanism against malignant transformation of cells; however, representatives of this species rarely get cancer.
To find out the mechanism of telomere length maintenance in bats, the scientists performed a comparative genomic analysis of the greater mouse-eared bat and about fifty other mammals, and of activation levels of over two hundred genes connected to telomere function. It turned out that the greater mouse-eared bat owes its long telomeres to about twenty genes. Among them is the SETX gene, coding an enzyme called helicase, which splits the double helix of DNA. There is also the ATM gene, which codes an enzyme called proteinkinase: when double-strand breaks occur, this enzyme activates proteins that stop the cell cycle and launch DNA reparation or apoptosis.
Incidentally, in humans, defects in these two genes cause various ataxias – a family of neurodegenerative disorders, where the underlying mechanism deals with accelerated telomere shortening. Therefore, it is quite possible that the proteins these two genes code do in fact help to maintain the stability of telomeres in long-lived bats
The record among rodents is held by the naked mole-rat, with its unique and scrupulously documented peculiarities of activation of a number of genes, DNA repair systems, etc. However, keep in mind that mole-rats have a very peculiar form of “social defense” of their colonies, with all individuals divided into “casts”. Many of the colony dwellers may end up in a snake’s bowels very quickly, thus allowing others to live extremely long lives – over 30 years, which is an absolute record for small rodents. When conducting research on the organization and functioning of the genome and cells of the naked mole-rat (Evdokimov et al., 2018), it would be extremely interesting and fruitful to compare this species not only with the house mouse, but with other animals which are considerably closer, for instance, with the African mole rats (Cryptomys), whose lives are significantly shorter – around 11 to 15 years.
Porcupines with their quills live long lives, as well as tree-dwelling squirrels and beavers barricading in dams and lodges. Small mouse-like rodents, who are the most numerous group of mammals both by species number and the number of individuals, live very short lives – only 2 to 4 years.
What about us? Tupaias, who are close to the ancient ancestors of primates, and prosimians have much shorter lifespans than true simians. The latter can get as old as 40 years. Apes, including gibbons, live relatively long lives – around 60 years. Apparently, this is the maximum biological age for humans, keeping in mind that the latest estimates put our median life expectancy at 38 years (Mayne et al., 2019). The following sixty years of human life, until its registered record of 120 years, are essentially “superbiological” and are mediated by changes in our social structure, medical advances, etc.

We are actively exterminating both ourselves and everything around us. Since 1900, humans have wiped out over 40 percent of all amphibian species, one third of species of the “immortal” corals, and over a third of all species of marine mammals. From the XVI century alone, we caused the extinction of 680 species of vertebrates, including 10 % of domesticated animal breeds, which had previously been used for food production. At least a thousand animal breeds are now on the verge of extinction.
Therefore, having reduced the number of our natural enemies nearly to zero, humans have acquired a new powerful enemy – that is, themselves. It is time for negotiations – for one thing, we must decide whether there is any good in extending our own lifespan beyond the share given to us by Mother Nature
Whether the latter is a good thing is very doubtful. Nature has erected mighty barriers against the biologically useless longevity, with cancer being the most important one. It greatly limits any attempts at “rejuvenating” and “improving” the genome. If one puts stem cells into tissues that have run out of their potential or edit some genes improperly – nature will answer with cancerous disease. One should not tune a working machine with a sledgehammer – and break what one did not build.
But there are no limits to the human curiosity and urge to act. No, we are not like that. So… you are saying jellyfish are immortal? What if we sink a barge with chlorine where they live, and spill a tanker of crude oil on the surface?
 But as long as mankind has spared some biological objects for its own scientifically minded fraction to study and compare, the following conclusions come to mind, at least in regard to vertebrates. Longevity is the combined attempts of some species and groups to greatly reduce the number of their natural enemies. For this purpose, terrestrial ancestors of whales, manatees and pinnipeds headed for the ocean; birds and bats flew into the sky. Many species, such as sharks, bears, or hippos, became super-dangerous themselves, or climbed high into trees, like primates and squirrels, or hid in inaccessible caves and holes, like the olm and the naked mole-rat, or just pretended to be piece of old rag, like the sloth. Therefore, increased lifespan is a survival strategy. But, apparently, it’s not the best one out there.
But as long as mankind has spared some biological objects for its own scientifically minded fraction to study and compare, the following conclusions come to mind, at least in regard to vertebrates. Longevity is the combined attempts of some species and groups to greatly reduce the number of their natural enemies. For this purpose, terrestrial ancestors of whales, manatees and pinnipeds headed for the ocean; birds and bats flew into the sky. Many species, such as sharks, bears, or hippos, became super-dangerous themselves, or climbed high into trees, like primates and squirrels, or hid in inaccessible caves and holes, like the olm and the naked mole-rat, or just pretended to be piece of old rag, like the sloth. Therefore, increased lifespan is a survival strategy. But, apparently, it’s not the best one out there.
Small rodents, such as mice and voles, the most numerous group among the mammals that has essentially conquered the world, are in general very short-lived. To comprehend the essence of their evolutionary success, one should compare the number of individuals and range of the trivial house mouse – so defenseless, vulnerable to all sorts of cancer, and short-lived, to those of the naked mole-rat. Human is thexonly vertebrate species that is trying to embrace both of the survival strategies: to be the innumerable frail house mouse and the immortal rare mole-rat at once.
References
Ackermann M., Stearns S. C., Jenal U. Senescence in a bacterium with asymmetric division // Science. 2003. V. 300. N. 5627. P. 1920.
Clay L. and Barral Y. New approaches to an age-old problem // Curr. Opin. Biotechnol. 2013. V. 24. P. 784–789.
Coelho M., Dereli A., Haese A. et al. Fission yeast does not age under favorable conditions, but does so after stress // Curr. Biol. 2013. V. 23(19). P. 1844–1852.
Evdokimov A. N., Kutuzov M. M., Petruseva I. O. et al. Naked mole rat cells display more efficient excision repair than mouse cells // Aging-US. 2018. V. 10. N. 6. P. 1454–1473.
Han S., Brunet A. Histone methylation makes its mark on longevity // Trends Cell Biol. 2012. V. 22(1). P. 42–49.
Lewin H. A., Robinson G. E., Kress W. J. et al. Earth BioGenome Project: Sequencing life for the future of life // PNAS. 2018. V. 115. P. 4325–4333.
Lind A., Lai Y. Y., Mostovoy Y. et al. A high-resolution, chromosome-assigned Komodo dragon genome reveals adaptations in the cardiovascular, muscular, and chemosensory systems of monitor lizards // Nat. Ecol. Evol. 2019. V. 3(8). P. 1241–1252.
Moskalev A. А., Kudryavtseva A. V., Graphodatsky A. S. et al. De novo assembling and primary analysis of genome and transcriptome of gray whale Eschrichtiusrobustus //BMC Evol. Biol. 2017. V. 17(258). DOI: 10.1186/s12862-017-1103-z.











