Why Do We Need to Read the Synedra acus Genome
In 2015, our research team (Limnological Institute, Siberian Branch, Russian Academy of Sciences) published the whole genome sequence of the diatom Synedra acus, which lives in Lake Baikal. Although now it is easier and much less expensive to “read” the genome of a living organism than a decade ago, this procedure still requires complex equipment and expensive reagents. So, why did researchers from the Limnological Institute tackle this large-scale project and choose this particular diatom as an object for their study?
Diatoms are of great interest to researchers working in most diverse areas of science. Owing to their abundance, diversity, and wide distribution—from oceans and lakes to peat bogs and Antarctic ice—diatoms account for up to 20% of the primary production in the biosphere; besides, these algal cells are enclosed in a wall of silica with an intricate nanosize structure, well preserved in geological deposits. However, the genomes of only six diatom species have been published, including Synedra, which is not enough for such a large group (Armbrust et al., 2004; Bowler et al., 2008; Tanaka et al., 2015; Galachyants et al., 2015).
For example, consider oomycetes, fungus-like organisms with multinuclear mycelium, which are closely related to diatoms: full-genome sequencing has been completed for four species of only one genus, Phytophthora, out of a hundred. The point is that Phytophthora species are notorious pathogens of cultivated plants; if the analysis of their genome makes it possible to design methods for their more efficient control, the money spent will immediately pay hundredfold. As for diatoms, they mainly interested basic science until recently. On the other hand, a successful “reconstruction” of the mechanisms underlying the formation of silica shells will be a real breakthrough in nanotechnologies when the necessary research is completed. Currently, fewer than ten laboratories worldwide are involved in diatom genomics.
Genome as a Key to the Evolution of Diatoms
Genome sequencing helps to solve several important problems that cannot be solved by other research approaches, such as microscopy or biochemical analysis.
The molecular clock approach has shown that diatoms emerged approximately 230 million years ago. Presumably, the first diatoms were naked flagellate cells; they are currently affiliated with Heterokonta because at some stages of their life cycle they possess a flagellum with finest bristles.
The earliest diatom deposits were found in onshore sediments in Korea. It was hypothesized that the ocean, having stepped back some 230 million years ago, left puddles of water inhabited by flagellate diatoms. They began to use silica, which retards aging, in order to survive the adverse conditions at rest.
Ancient diatoms consumed so much silica that it started to accumulate in cells, possibly as small scales at the initial stage; the scales were forced out of the cells, forming a protective envelope around them, which prevented dehydration when shallow water bodies dried (according to Medlin and Sims, 2006)
First, genetic information considerably enhances the search for proteins responsible for particular functions (Galachyants et al., 2015). Once the genome project is completed, several tens of thousands of “read” genes and, correspondingly, predicted proteins are at the disposal of the researcher. Part of these genes is recognizable right away because analogous genes have been already studied and described in other organisms. As for the remaining genes, it is at least possible to estimate the molecular weights of the proteins they encode and search for their “relatives” in other genomes. It is also sufficiently easy to find out whether the protein of interest is a transmembrane protein or not, and to which cell organelle it is delivered after synthesis. Moreover, if transcriptome (the total set of all nucleotide sequences “read” from the genes, including mRNA and noncoding RNA, synthesized during a particular time interval) data are available, it is possible to clarify the conditions when this or that gene is expressed, or starts to work.
First and foremost, our team has focused on the genes involved in silicon metabolism, while the goal of Tanaka’s research team, which sequenced the genome of Fistulifera solaris, was utilization of this marine diatom as a biofuel producer (Tanaka et al., 2015).
Second, only if complete genomes are available, it is possible to study certain aspects of evolution; note that the evolution of heterocont algae, which include diatoms, is most intriguing.
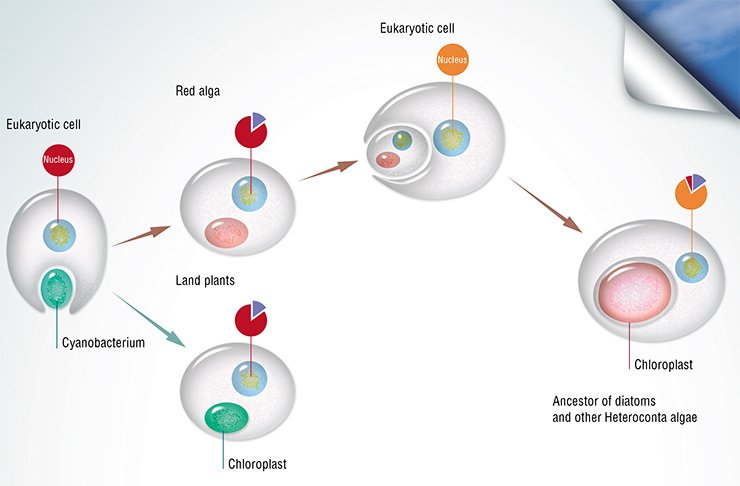
It is currently regarded as proved that plant chloroplasts, in which photosynthesis takes place, originated via endosymbiosis: once a cyanobacterium cell was taken up by a eukaryotic cell, but was not digested; it started to function within the host, gradually reducing to the current state of an intracellular organelle. In this process, the endosymbiont donated most of its genes to the host genome. As for heteroconts, and in particular diatoms, it is postulated that their ancestors, in turn, “swallowed” a eukaryotic red alga together with its chloroplast. This theory is confirmed by the presence of four (instead of commonly met two) membranes surrounding the chloroplasts of diatoms as well as over 170 genes of red algae discovered in their genome (Bowler et al., 2008). That is why diatom genomes contain genes traced back to the participants of both endosymbiotic events. Diatom genomes also include genes belonging to various prokaryotes (proteobacteria, archaea, etc.), brought there by the so-called horizontal transfer, as well as numerous unique genes that have no analogs in any other living organisms.
To trace the history of an individual gene, it is sufficient to sequence this gene alone for a certain sample of organisms, which is considerably faster and less expensive as compared with whole-genome sequencing; besides, many more species can be analyzed. However, the reconstruction of a complex evolutionary event requires studying the histories of all genes in all representatives of the group; this information can only be obtained through whole-genome studies.
In our case, the selection of an object for research was due to the fact that Synedra acus is a freshwater species. The fact that this diatom is able to extract the necessary amount of silicon even from the low- mineralized Baikal water makes Synedra an ideal candidate for studying the mechanism of silicon import as well as for learning about other adaptations to freshwater life. Second, this alga has become the first representative of a large group of araphid pennate diatoms, which makes it possible to track the history of development of certain molecular mechanisms and of specific genome features among different diatom taxa.
Our work on the Synedra acus genome does not end with the publication of its genome sequence. We have already obtained promising data on the structure of individual genes involved in the silicon metabolism, and the study of corresponding proteins is in progress. We intend to obtain transcriptome, which will help to assess the expression of target genes and to gain a deeper insight into their regulation.
References
Armbrust, E. V. et al. The Genome of the Diatom Thalassiosira pseudonana: Ecology, Evolution and Metabolism // Science. 2004. Vol. 306 (5693). P. 79—86.
Bhattacharya, D., Qui, H., Price, D. C. Why we need more algal genomes. // J. Phycol. 2015. Vol. 51 N.(5). P. 1—5.
Bowler, C. et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes //Nature. 2008. 456: 239—244.
Galachyants, Yu. P. et al. Whole genome sequencing of the araphid pennate diatom Synedra acus subsp. Radians from Lake Baikal. Dokl. Ross. Akad. Nauk. 2015. V. 461. N. 3. P. 348—352.
Tanaka, T. et al. Oil accumulation by the Oleaginous Diatom Fistulifera Solaris as Revealed by the Genome and Transcriptome // The Plant Cell. 2015. Vol. 27 N.(1). P. 162—176.