Genetic Alphabet: Expanded or Not?
The top ten breakthroughs of 2014 include a study by Floyd Romesberg and his group at the Scripps Research Institute (La Jolla, United States), who not only made DNA with two new letters of the genetic code but also tricked it to replicate in Escherichia coli, a workhorse of molecular biology
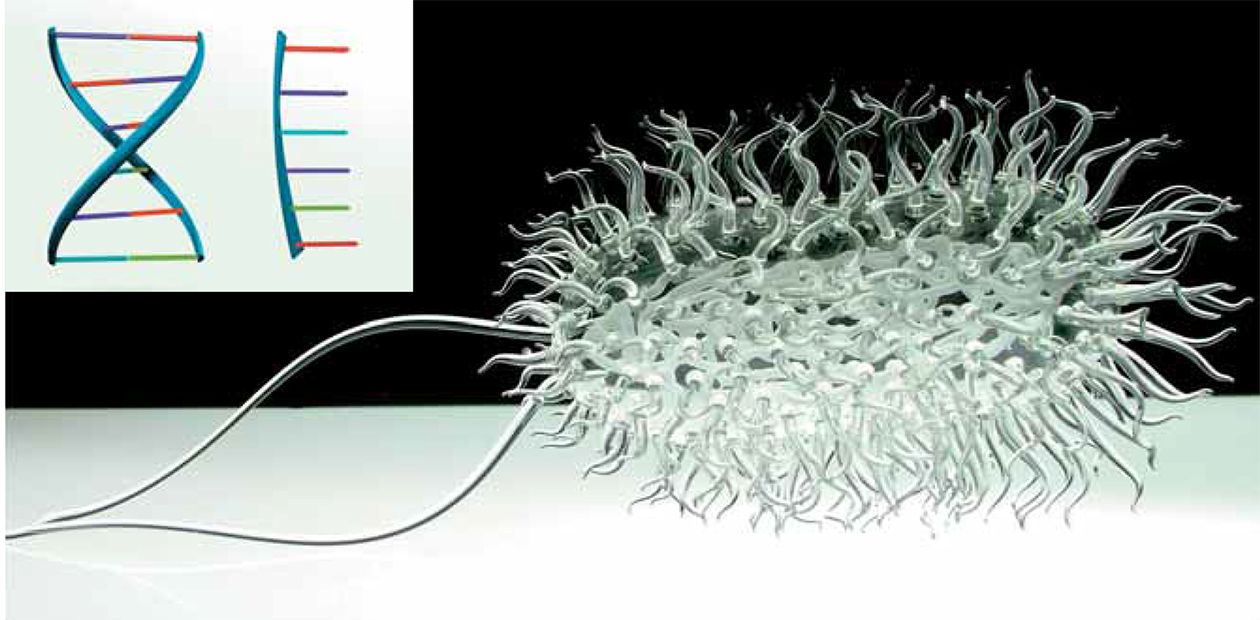
From school biology lessons, everybody knows that the text of the DNA molecule—the storage of genetic information—is written with four letters only: A, T, G, and C. These letters stand for four nucleobases—adenine, thymine, guanine, and cytosine—which are attached to the sugar-phosphate backbone. The two resulting strands form the well-known DNA double helix, which contains the blueprint for building and running our bodies.
The two DNA strands are complementary: A in one strand always stands opposite to T in the other strand, and G is always opposite to C. The “opposite” bases in the pairs are connected by hydrogen bonds, which ensure the matching of the bases in a pair. For instance, no hydrogen bonds can form between A and C; thus, the complementary bases fit like a lock and key.
When the structure of DNA was discovered in the mid-20th century, scientists began to wonder why Nature had chosen these two base pairs and not others, and whether it is possible to tamper with the cell so that it would use any alternatives? This question is still unanswered. Many researchers in the field of prebiotic evolution believe that this choice was made purely by chance and took hold later, after passing through the most difficult stage in the emergence of life on our planet—when molecules became replicators, i.e., learned to reproduce themselves.
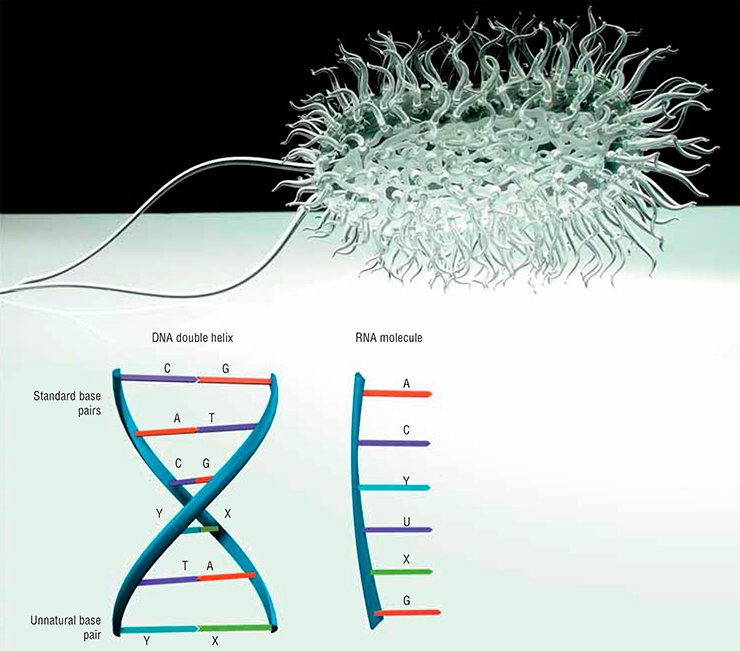 The probability of this event is very low. Not surprisingly, even if there had been complementary pairs involving other bases, they would have passed through this bottleneck. However, it is most likely that the first replicator molecule was RNA, not DNA. RNA uses, instead of thymine, another nucleobase—uracil, which also forms pairs with adenine. In the transition to the DNA world, uracil was replaced by thymine for reasons related to the reliability of information storage.
The probability of this event is very low. Not surprisingly, even if there had been complementary pairs involving other bases, they would have passed through this bottleneck. However, it is most likely that the first replicator molecule was RNA, not DNA. RNA uses, instead of thymine, another nucleobase—uracil, which also forms pairs with adenine. In the transition to the DNA world, uracil was replaced by thymine for reasons related to the reliability of information storage.
Curiously, we already know organisms in which the Great Four differs from the one described in textbooks. For instance, the DNA of many bacteriophages (viruses that infect bacteria) contains no thymine, which is replaced by uracil, hydroxymethyluracil, or other uracil derivatives with an additional attachment of a sugar residue. This substitution helps the virus protect itself against bacterial defense systems, which eliminate alien DNA. Moreover, in the 1970s, biologists from Leningrad found in a most ordinary puddle a bacteriophage whose adenine was completely replaced by another base, 2,6-diaminopurine.
The second of the questions posed above triggered the development of a new field of molecular biology—the creating of an expanded genetic code. Researchers working in this field not only seek to make alternative base pairs but also look for ways to incorporate unnatural amino acids into the structure of proteins (the genetic systems of all currently known living organisms are known to code for exactly 20 standard amino acids). Clearly, if we learn how to assemble DNA from an expanded repertoire of base pairs and equip the code with the ability to incorporate nonstandard amino acids into proteins, we will open up unprecedented new opportunities for synthetic biology, a field of science that deals with the creation of living systems and processes that do not exist in nature.
Compared with such a global problem, the breakthrough reported by Science does not look as a stunning discovery, but rather as the next stepping stone (and not a very big one) on the path researchers set out on two decades ago. The main conceptual breakthrough on this path was made in the late 1990s by a research team led by Eric Kool (University of Rochester, United States), who showed that hydrogen bonds are not needed at all to make a stable base pair that fits well into the DNA double helix. One can create artificial bases containing no atoms capable of forming these bonds whatsoever, and they will be able not only to stably exist in DNA but also to be incorporated into DNA by ordinary DNA polymerase enzymes (at least by some of them).
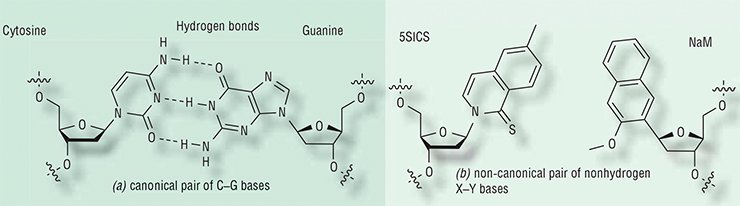 Unnatural bases have been also studied for many years at Floyd Romesberg’s laboratory, whose results attracted the attention of Science. However, until recently all the studies were conducted in vitro, not in a living cell. This time Romesberg’s team took one of these nonhydrogen base pairs and tried to force it to replicate inside E. coli bacteria, which are traditionally used in molecular biology experiments.
Unnatural bases have been also studied for many years at Floyd Romesberg’s laboratory, whose results attracted the attention of Science. However, until recently all the studies were conducted in vitro, not in a living cell. This time Romesberg’s team took one of these nonhydrogen base pairs and tried to force it to replicate inside E. coli bacteria, which are traditionally used in molecular biology experiments.
In a living organism, however, bases do not appear at the will of the experimenter. Behind each of the four DNA letters, there is a multistep pathway for their synthesis in the cell, and the latter, of course, cannot make the unnatural bases invented by chemists. Therefore, the scientists cheated: they expressed a protein from the cell wall of the diatomic alga Phaeodactylum tricornutum inside the bacteria, because this protein can capture individual DNA letters directly from the environment. Consequently, unnatural bases (more exactly, not the bases themselves, but deoxynucleoside triphosphates, i.e., building blocks with a part of the sugar–phosphate backbone, which combine to form DNA) were simply added to the culture medium in which these bacteria were growing.
However, there was one more problem to be resolved. Bacterial cells cannot use unnatural bases in large quantities—the bacteria would not survive because the existing genetic apparatus would not recognize them. Therefore, the scientists put only one unnatural pair and not even directly into the bacterial genome but into a plasmid, a small circular molecule of the DNA, which can independently exist and replicate inside the bacterial cell. And since the DNA polymerase III enzyme, which is responsible for the bulk replication in bacteria, does not recognize nonstandard bases at all, the scientists had to put the unnatural base pair not simply into the plasmid, but into a small segment of it, which is synthesized by another enzyme, DNA polymerase I.
After all these manipulations, bacteria were grown in a medium containing nonstandard bases for 15 hours; during this time the cells divided 24 times. Then the researchers checked what was in that place of the plasmid where they had put the unnatural pair. If the cell had been unable to use the matching unnatural nucleotides for replication and incorporated normal ones opposite to them, the unnatural pair would have been retained after 24 divisions only in one plasmid out of 17 million copies. But nothing like that happened: the unnatural pair was retained in 86% of the plasmid copies and was eventually lost only after several days of further growth.
No one can deny the importance of the work done by Romesberg’s team: they have been the first to show that an unnatural base pair can function in a living cell. But it is too early to say that they have generated an organism with “an expanded genetic alphabet.” Ostensibly, Nature somewhat overhyped the paper’s title. The authors of the article bypassed the major unresolved problems of artificial genetic code. The actual expansion of the DNA alphabet requires that researchers should at least integrate the synthesis of unnatural nucleotides into the cell, make these nucleotides compatible with the general replication system, and, most importantly, figure out how to use the new letters to make the cell produce new proteins.
The task still looks extremely complicated—like a space flight in the early days of aeronautics. In this sense, the work done by Romesberg’s team can be compared with the launch of a hot air balloon by the Montgolfier brothers. But it was not balloons that flew eventually into space. Thus, although the breakthrough reported by Science is certainly a step in the right direction, it is unclear whether this path will lead sinthetic biology to its ultimate destination.
References
Vlassov V. V. and Vorob’ev P. E. RNA world: yeasterday and now // Science First Hand. 2015. No. 1 (40). pp. 6—15.
Malyshev D. A., Dhami K., Lavergne T., et al. A semi-synthetic organism with an expanded genetic alphabet // Nature. 2014. V. 509. N. 7500. P. 385—388.
Translated by A. Kobkova