To Help a Structural Biologist
X-ray structure analysis has come to stay in the life of biologists since the mid- 20th century. It was at that time when John Kendrew from the famous Laboratory of Molecular Biology in Cambridge succeeded, after many years of work, in determining the structure of a protein, which was sperm whale myoglobin, assisting the whale to store oxygen in its muscles when diving. Max Perutz, a colleague of Kendrew who later shared a Nobel Prize with him, at the same time determined the structure of the protein hemoglobin, familiar to everybody, which transfers oxygen in the blood. Just note that Perutz, who started this work at the age of 25, completed it only 22 years later!
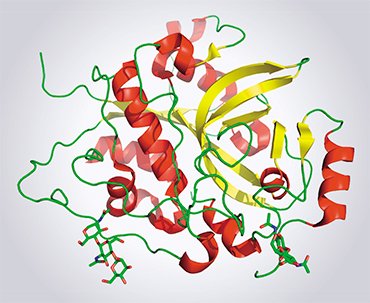
Currently, X-ray structure analysis is one of the two approaches allowing the structure of most complex molecules, such as proteins, to be solved with an atomic resolution (the second approach is nuclear magnetic resonance spectroscopy). In an ideal case, this method makes it possible to determine the coordinates of each atom in a molecule except for hydrogen.

A notable downside of this method is its exclusive labor intensity, although the NMR approach it isn’t any better in this respect. Despite all the improvements introduced since the time of Perutz and Kendrew, it has three bottlenecks limiting its throughput. First, X-ray structure analysis requires rather large protein crystals, which will produce a high-quality pattern of X-ray diffraction on the atoms in protein molecules ordered in a crystal lattice. Protein crystallization is still black magic in many respects, impossible to achieve for some proteins, and the resulting diffraction pattern is not always amenable to analysis.
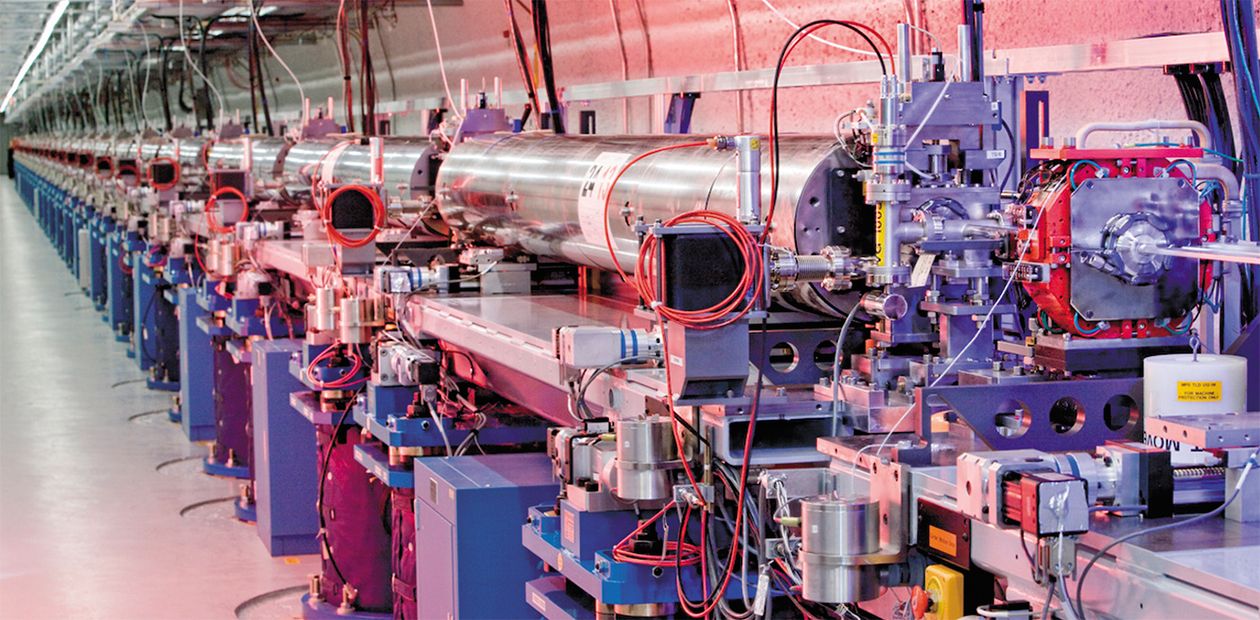
Second, even with a good protein crystal, taking a diffraction picture requires a quite bright (that is, high power) beam of X-ray radiation. Currently, the beams are generated in large synchrotrons with the help of specialized hardware, and the waiting lists for such stations are months and even years long.
 The third constraint, the time it takes to decode the protein structure from a diffraction pattern, has become less crucial with the advent of powerful computers and improved algorithms, but the first two issues until recently have remained serious obstacles. However, the award for their overcoming is high; for example, knowing the structure of a protein involved in pathological processes, it is possible to design the drugs that may bring billions in sales. That’s why researchers from pharmaceutical companies spend day and night at stations with synchrotrons paying a pretty penny for this.
The third constraint, the time it takes to decode the protein structure from a diffraction pattern, has become less crucial with the advent of powerful computers and improved algorithms, but the first two issues until recently have remained serious obstacles. However, the award for their overcoming is high; for example, knowing the structure of a protein involved in pathological processes, it is possible to design the drugs that may bring billions in sales. That’s why researchers from pharmaceutical companies spend day and night at stations with synchrotrons paying a pretty penny for this.
For a good while, the motto for the improvement of X-ray structure analysis has been “Smaller, brighter, faster”: it is easier to grow a smaller crystal, and brighter radiation sources allow faster data recording. The breakthrough of the year reported by the Science magazine raised the bar in this competition very high.
A superpowerful X-ray laser at the Stanford Linear Accelerator Center in California shoots pulses 40 femtoseconds long (that is, 40 quadrillionth fractions of second – light can pass a distance of only 12 micrometers for this time!). The radiation brightness is so high that it allows 10- to 100-fold smaller crystals to be used for imaging. The laser pulse literally evaporates the crystal, but the diffraction pattern can be recorded. Several hundreds of such snapshots give the pattern whose quality is not lower than that of commonly recorded ones; however, the former is obtained over billionth parts of a second.
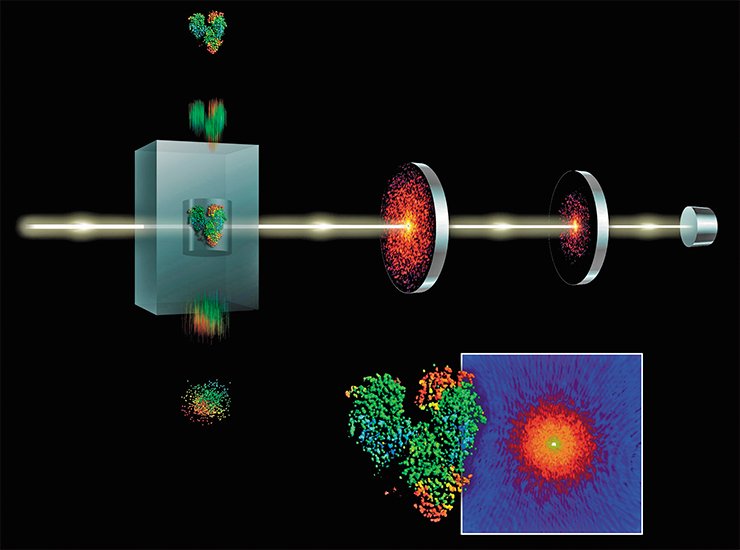
So far, only one protein structure has been determined using this unique laser. This protein is cathepsin D, an important cell component of the parasite trypanosome (Trypanosoma brucei), the agent causing sleeping sickness. As for structural biologists, they wonder whether the X-ray lasers have a promising future in this field. Indeed, despite the dazzling results, such a station is much more complex and expensive as compared with the conventional stations with synchrotrons, whose number reaches several dozens worldwide.
However, it is hoped that the X-ray laser might make a lifelong dream of biologists come true, allowing them to record a diffraction pattern from individual protein molecules without any crystallization at all. This will open yet unimaginable prospects for researchers.