Taming the Ancient Molecule
RNAs have always been in the spotlight; scientists working for the Novosibirsk Institute of Chemical Biology and Fundamental Medicine had been interested in RNAs even before this research institute was founded. As early as the 1960s, when the future institute was the Department of Biochemistry with the Novosibirsk Institute of Organic Chemistry (Siberian Branch, USSR Academy of Sciences), Academician Dmitry G. Knorre, head of department, and his colleagues formulated the concept of complementarily addressed modification, that is, the “targeting” of short nucleic acid molecules (oligonucleotides) carrying active chemical groups to RNA molecules based on the principle of complementarity. Researchers of the Institute of Chemical Biology and Fundamental Medicine have significantly contributed to the research into transfer RNA and ribosomes, to the verification of the hypothesis on “RNA world,” and to the elaboration of new methods for the chemical synthesis of RNA molecules. The catalytic RNAs—ribozymes—were not overlooked either. That is why the proposal to organize at the institute a laboratory led by the Nobel Prize laureate who discovered the catalytic properties of RNA seemed natural; it was supported by the Ministry of Education and Science, which awarded one of its few and highly prestigious “megagrants” for this purpose
In the beginning was the word of three letters. And this word was RNA…
When Sydney Altman started to study the enzyme RNase P, which eventually brought him the Nobel Prize, there was a unanimous opinion in biology that all the catalysts in the cell were proteins. This opinion was confirmed by the Nobel Prize (1946) awarded to the American biochemists James Sumner, John Northrop, and Wendell Stanley. This was a milestone event: the American trio had demonstrated that enzymes were biological catalysts, that is, they represented certain chemical compounds. This finally buried the doctrine of vitalism, “the vital force,” running like a golden thread through the fabric of the pre-20th century biology.
Since then, the protein nature of enzymes has not been doubted. This is why Altman and his colleague Thomas Cech, who discovered a catalytic role of RNA in excising noncoding regions (introns) from itself, had to put in great efforts to publish their results, not fitting in the paradigm of that time. However, what distinguishes true science is that most unbelievable results soon become commonly accepted facts if other scientists can reproduce them. Cech and Altman gained their credit very soon: their pioneer papers appeared in 1982 and 1983, and they bagged the prize in 1989, which is almost instantly, as times go.
The work by Altman and his colleagues caused a surge of interest in RNA. Another indestructible paradigm of that time—the so-called “central dogma of molecular biology”—stated that RNA (namely, messenger RNA, mRNA) carried information between DNA and proteins. Noncoding RNAs—ribosomal and transfer, being major players in translation (protein synthesis according to the enzyme template)—were also well known.
It was believed that the RNA functions ended up there. True, short nuclear RNAs with unknown functions were already known, but hardly anybody at that time expected any surprises from RNA. However, Francis Crick, the creator of the central dogma, and some other prominent scientists involved in the research into the origin of life (Leslie Orgel and Carl Woese) hypothesized that at the dawn of life RNA could have played a much more important part than nowadays. However, these theories were based on most indirect considerations, for example, on the idea that many protein enzymes function only in the presence of “assistants,” coenzymes of a nucleotide nature, which, presumably, are “fossil remains” of the ultimate ancestral forms of living beings.
The discovery of RNA enzymes (hereinafter, referred to as “ribozymes,” as is commonly accepted) changed the vision of what this molecule could and could not do. In 1986, Walter Gilbert, another Nobel Prize winner, for the first time coined the term “RNA world,” i.e., that is, our world at the very early stages of its evolution when there were neither DNA nor proteins, and only self-reproducing catalytic RNA molecules competed with each other for “survival.”
Since then, ever more new proofs for this challenging hypothesis have appeared. The number of the known ribozymes, both natural and artificial, has reached several hundred and many new biological systems have revealed the role played by RNAs. Just a curious fact: in 1998, a rock band named Ribozyme appeared in Norway.
RNA here, RNA there
However, the ribozyme discovery alone would be insufficient for the RNA to turn from a modest intermediate link between the major carrier of genetic information, DNA, and the major executors of cell functions, proteins, into the central element uniting a great many vital cellular processes. After all, the number of ribozymes is small as compared with the number of proteins and their functions in nature are rather limited. However, since the discovery made by Altman and Cech, RNA has sprung surprises on us almost every year!
So what new functions of this molecule we have come to know beyond the simple scheme by Crick, which comprises only the messenger, ribosomal, and transfer RNAs? We can start from this very scheme: it has turned out that in such a complex structure as the ribosome (which even in bacteria consists of three strands of different rRNAs and several tens of proteins), the major reaction, namely, the peptide bond formation, is catalyzed not by the proteins but by an RNA molecule!
Today, we know many situations when RNA acts as a catalyst in a complex with proteins or drives the assembly of a functional RNA–protein complex. One nifty and important example of such a complex is the enzyme telomerase, first predicted with the point of his pen by Alexey Olovnikov, a Soviet scientist, and then discovered by American biologists Elizabeth Blackburn and Carol Greider, who won a Nobel Prize for this work.
The chromosomes of humans and all other eukaryotes are linear, and replication of the ends in linear DNA presents a special problem: because of specific features of the mechanism initiating DNA synthesis, with each cell division the terminal DNA regions are lost. However, a specific structure of the chromosome tips (telomeres) gets around this problem.
Replication of the ciliate Tetrahymena was first studied by Blackburn and Greider; this protozoan contains many thousands of linear microchromosomes in its somatic nucleus. The telomeres consist of numerous conserved short (hexamer) repeats, TTAGGG in human and TTGGGG in Tetrahymena. The actively dividing cells contain a specialized enzyme involved in the replication of the terminal DNA sequence, referred to as telomerase; the infusorian telomerase contains a small RNA molecule which carries the sequence AACCCCAAC.
It is evident that this sequence is complementary to the complete hexameric repeat plus its half. Telomerase binds to the DNA end; synthesizes six nucleotides, GGGTTG, using its RNA as a template; and then shifts by six nucleotides to repeat this cycle. In this case, the protein moiety of telomerase acts as an enzyme, while RNA is utilized as a template for synthesis.
Such functional ribonucleoproteins are ample in the cell. The process of splicing (excision of introns from mRNA) is mentioned above when discussing Cech’s discovery of ribozymes. Interestingly, he discovered the ribozymes involved in splicing in the very same ciliate Tetrahymena, which thus made history in molecular biology.
Anyway, splicing in most of the organisms, humans included, is catalyzed by small nuclear ribonucleic particles rather than ribozymes in a strict sense; these particles are RNA–protein complexes where RNA plays both the structural and catalytic roles. Another, rather unexpected, example is that the proteins secreted from the cell have to pass through the endoplasmic reticulum membrane during the synthesis; for this purpose, they interact with the so-called signal recognition particle (SRP), a ribonucleoprotein comprising one RNA molecule and six proteins.
In the early 2000s, the RNA switches of biochemical processes, or riboswitches, were discovered. These regions of mRNA molecules are noncoding but able to bind certain signaling molecules in the cell, thereby changing their structure and influencing the course of transcription. In particular, the thiamine riboswitch, discovered by Russian scientists from the Institute of Genetics and Breeding of Industrial Microorganisms (Moscow), binds to thiamine pyrophosphate, a derivative of the B1 vitamin, which decreases mRNA translation of the genes involved in the synthesis of this vitamin. Once this vitamin is present in large amounts, the cell may shut down its production.
Quite frequently, we know that a noncoding RNA is involved in some process but we cannot determine the mechanism by which it proceeds. For example, one of the two X chromosomes in all the cells of female mammals is condensed and functionally inactive. This condensation requires a noncoding RNA, Xist, multiple copies of which cover one of the X chromosomes. It is still far from clear how Xist RNA binds to the chromosome and which of the two chromosomes are selected. Here in Novosibirsk, a team led by Suren Zakian (Institute of Cytology and Genetics, Siberian Branch, Russian Academy of Sciences) contributed much to the research into the functions of this RNA.
 To start its replication (doubling of the genome), the human cell requires the so-called Y RNA, which binds to the replication origins. However, what goes on at a later time is still a riddle.
To start its replication (doubling of the genome), the human cell requires the so-called Y RNA, which binds to the replication origins. However, what goes on at a later time is still a riddle.
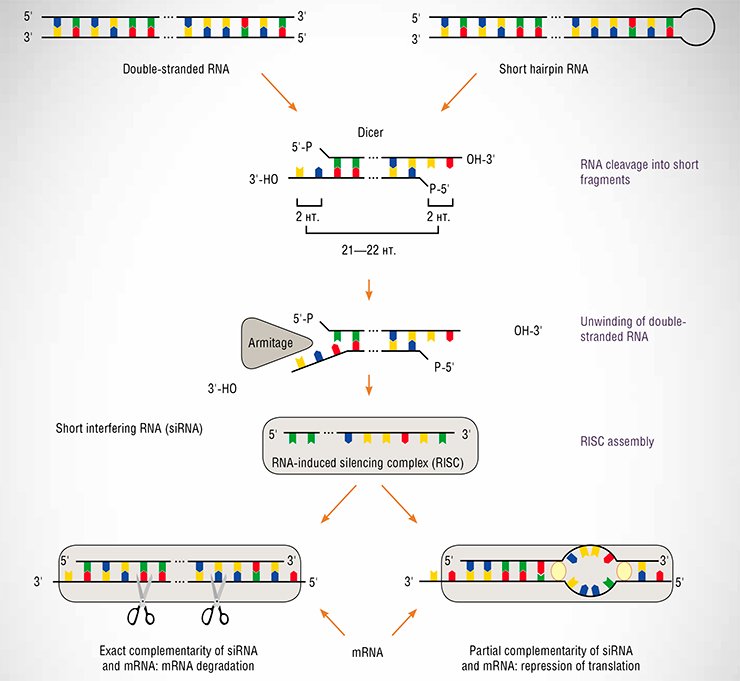
Finally, noncoding RNAs frequently act as gene activity regulators utilizing the complementarity principle; this idea, proposed by Academician Knorre as a human intervention into the cell affairs, turned out to be invented by Nature, too. Consider, for example, the phenomenon of RNA interference, discovered in 1998 by two American researchers Craig Mello and Andrew Fire. Actually, this phenomenon was observed in plants as early as the 1920s but was not correctly interpreted at that time. The essence of RNA interference is that linear double-stranded or hairpin RNAs can inhibit the expression of the genes carrying the sequences homologous to them. The efforts of many scientists have allowed the mechanism of this process to be clarified. As a rule, mRNA is degraded when it carries a fragment precisely complementary to short interfering RNAs (siRNAs) or its translation is merely blocked when the complementarity is partial. However, the molecular details of these processes are still under intense scrutiny. RNA interference can also regulate the chromatin structure and the degree of its condensation.
Once the RNA interference mechanism was clarified making it possible to inhibit the expression of almost any gene in the cell, researchers started to devise various useful applications for this tool. Of course, this started as a lab exercise: the gene expression inhibition with the help of RNA interference or gene knockdown (by the analogy to gene knockout, i.e., making a gene inoperative) came to stay in the molecular biological laboratory toolbox. The specialized services are now available that design the RNA sequence and make the expression construct to knock down a gene of your interest.
Naturally, the useful laboratory tool soon made its way intodesigning therapeutics involving RNA interference. The very first interfering RNAs tested in clinical trials were intended for the reversion of retinopathy and treatment of respiratory syncytial virus infection, a major respiratory disease in newborns and babies. This was not the end: antiviral RNAs against herpes, hepatitis A and B, AIDS, influenza, measles and even some cancers are now close to clinical trials.
RNA interference has even found its application in agriculture. In particular, Monsanto, a well-known biotechnology company, is now breeding the plant cultivars able to produce insecticide interfering RNAs. Another example is a cotton cultivar with edible seeds , in which RNA interference prevents synthesis of the toxin gossypol.
What remained unclear by the end of the last century was why the cell has a special mechanism that works with the RNA molecules introduced there by researchers. Surely cells have reasons to possess proteins that in a concerted manner use something that scientists push there?
Thus, a new discovery, microRNA, in the early 2000s was not quite unexpected. As usual, it appeared that the first RNA of this kind had been found earlier in a microscopic nematode, Caenorhabditis elegans, a model object for developmental genetics; however, the essence of this RNA type at that time was not revealed.
In the cell, microRNAs are generated from their precursor RNAs much in the same way as from the artificially introduced double-stranded RNAs, that is, first, the molecules are cut into pieces, and then fed into the RNA interference pathway. Hundreds of microRNAs have been discovered by now: presumably, they are among the most important regulators for human gene expression.
Russian nucleic acids
The goal of Altman’s laboratory in Novosibirskof, organized thanks to a megagrant, is far from being simple: the researchers aim to create an entirely new class of drugs to treat bacterial, viral, and protozoan infections. Currently, microbes have acquired resistance to many antibiotics and, facing the truth, it is clear that the current health care is losing the race: staphylococcus and tuberculosis strains resistant to almost all known antibiotics have emerged. On the other hand, the uncontrolled use of antibiotics not only fuels the evolution of drug resistance, but puts the patients themselves in danger, because antibiotics kill both pathogens and useful dwellers of our gut.
This suggested applying gene-targeted compounds to control pathogens, for such drugs will only kill the target microbe and will not affect the remaining bacteria. Once the structure of a bacterial gene is known, it is possible to synthesize an appropriate oligonucleotide—a fragment of RNA, DNA, or some modified nucleic acids—that will “stick” to the corresponding nucleic acid target. If this guidance part is equipped with a molecule able to destroy the target nucleic acid, we will have the gene-targeted reagent acting only on the target genome. Of course we still have to protect this drug from destruction while it is in the body, make sure it is not toxic, and, most importantly, deliver it inside the cell.
Collaborating under the megagrant on the Russian side are chemists, who synthesize nucleic acids, and microbiologists, who test these compounds using real targets. Some aims have already been reached: for example, a variant of the oligonucleotide that is not destroyed in blood has been designed.
As for the problem of drug delivery to bacterial cells, it is still to be solved. For this purpose, the experts from the Institute of Chemical Biology and Fundamental Medicine screen libraries of natural peptide carriers.
Concurrently, an oligonucleotide targeted against influenza virus is being designed; this is a relatively simple task, since the nucleic acids of the virus are in the infected human cell, which is much easier to enter than the bacterial cell, enclosed into a thick protein–polysaccharide capsule. Thus, Novosibirsk scientists follow many lines at the same time, and most likely, at least one of these variants will bring success.
New tricks of the tamed RNA
Another challenge accepted by the Siberian laboratory of the Nobel Prize winner is taming RNA for yet another purpose, cell therapy.
This project has its own intriguing and rich history. In a way, it starts from Dolly, the sheep cloned in 1996 from an udder cell by Ian Wilmut. This way of cloning, somatic cell nuclear transfer, requires an enucleated egg cell and a donor cell, which provides the genetic material for the embryo. At the earliest developmental stages, embryonic cells produced by such a transfer retain their pluripotency, i.e., the ability to develop into any cell of an adult body.
After the works of Wilmut, an ethically frightful prospect loomed over the humankind: an adult person seeking rejuvenation creates his/her own clone to extract either the cells, at an early stage of embryo development, or organs later in life. This fear is not groundless: modern medicine knows cases when parents deliberately gave birth to a healthy child to make a bone marrow donor for an elder sibling suffering from a rare hereditary disease.
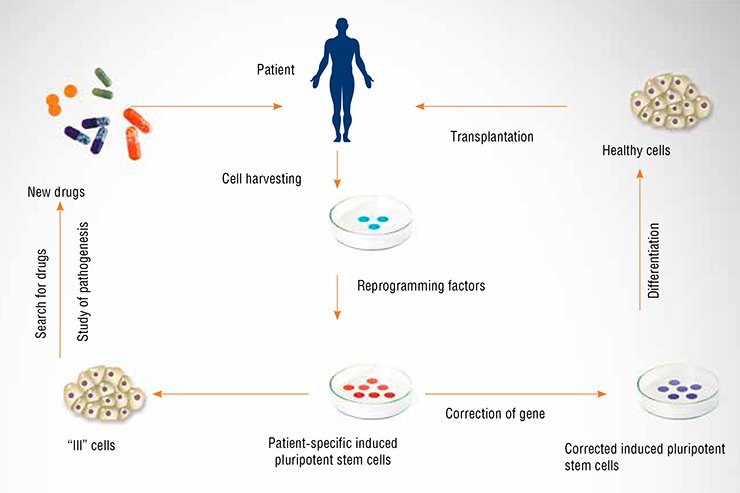
However, Shinya Yamanaka, a Japanese researcher, sliced the Gordian knot in 2006 by devising a method to obtain pluripotent cells from almost any body cell without the need to create and subsequently to destroy an embryo. It has turned out that such a transformation needs actively working copies of only four master regulator genes—Sox2, Oct4, Klf4, and c-Myc—to be introduced into the cell. As a result, a liver cell, for example, becomes an induced pluripotent stem cell (iPS cell), which can be further transformed into a liver cell, a neuron, or a leukocyte. Laboratory mouse models even showed that these cells could give rise to pups and then to healthy and fertile adult mice. The discovery by Yamanaka gave hope that we would soon learn how to heal almost any hereditary disease. Indeed, if it were possible to take cells of a patient, induce them to a pluripotent state, correct the corresponding genetic defect in these cells, and graft them back into the patient, this would rid the humankind of many genetically determined disorders.
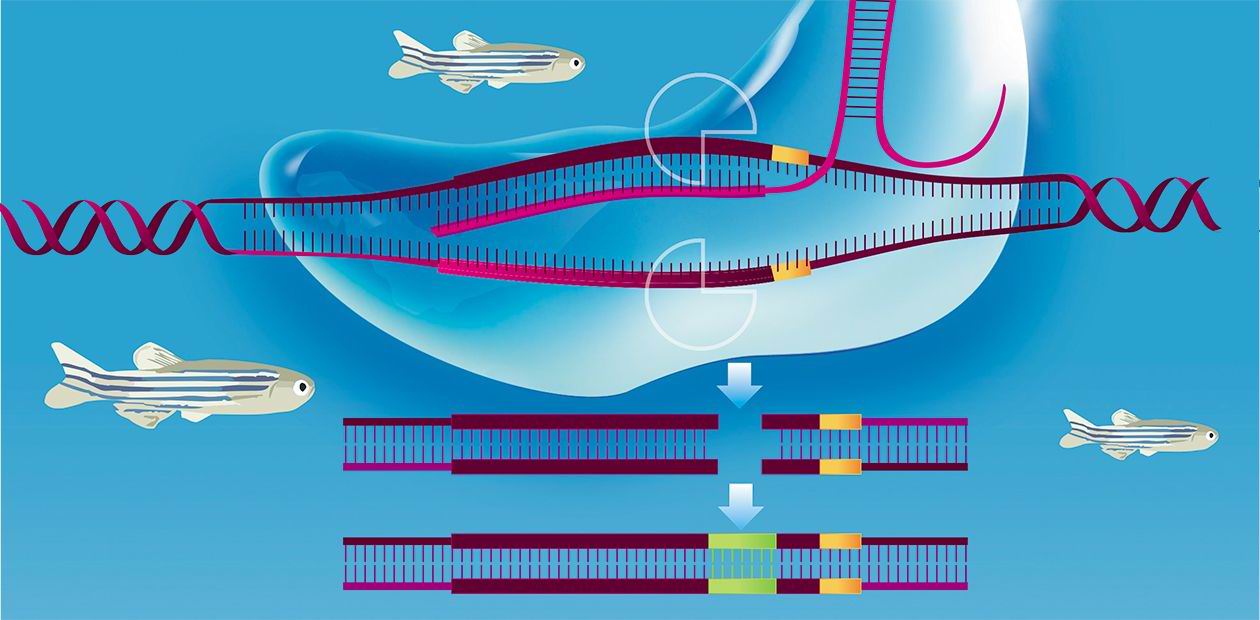
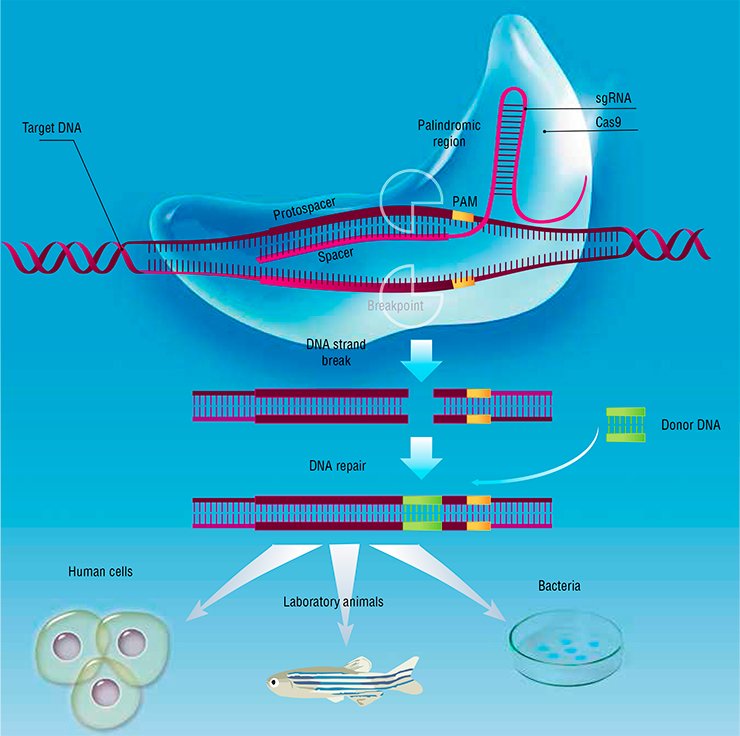
 All this is excellent, but how can we correct the cell genome? In principle, biologists can do it by homologous recombination (this is how transgenic animals have been produced since the late 1980s), but this procedure is blatantly inefficient. The solution came quite unexpectedly. In 1987, numerous clustered regularly interspaced short palindromic repeats (CRISPR) were discovered in the E. coli genome; they are used as a template for synthesizing small RNAs. It has emerged that bacteria use these repeats for defense against bacteriophages to recognize and destroy the intruder’s nucleic acids, dispatching small RNA fragments to bind them. These RNAs do not act alone but together with a protein, Cas9, which cleaves viral DNA or RNA.
All this is excellent, but how can we correct the cell genome? In principle, biologists can do it by homologous recombination (this is how transgenic animals have been produced since the late 1980s), but this procedure is blatantly inefficient. The solution came quite unexpectedly. In 1987, numerous clustered regularly interspaced short palindromic repeats (CRISPR) were discovered in the E. coli genome; they are used as a template for synthesizing small RNAs. It has emerged that bacteria use these repeats for defense against bacteriophages to recognize and destroy the intruder’s nucleic acids, dispatching small RNA fragments to bind them. These RNAs do not act alone but together with a protein, Cas9, which cleaves viral DNA or RNA.
Why not use this system against another, non-viral target? It is sufficient to devise the necessary guide RNA, and the CRISPR/Cas9 system will cleave any DNA we wish. Moreover, if the cell is supplied with a “donor” of the necessary genetic material (for example, an oligonucleotide with the corrected sequence), the break will be sealed by the cellular DNA repair systems. Thus, any mutation in the genome can be edited out. It is prudent to say that the CRISPR/Cas9 system is not the only currently available way to edit the genome, but easily the most efficient one.
Naturally, the Novosibirsk research team could not but pay attention to such a promising tool. They plan to produce the cells that carry mutations associated with the hereditary forms of Alzheimer’s, Parkinson’s, and some other diseases. Such cells are of a paramount importance for drug design, since only human cells can provide an adequate insight into human pathologies. Animal models are inapplicable in many cases—even the great apes never get Alzheimer’s disease. On the other hand, this system allows the cells of the patients with hereditary diseases to be converted into perfectly healthy cells. For this purpose, the team of the Institute of Chemical Biology and Fundamental Medicine in collaboration with the Meshalkin Institute of Circulation Pathology selected the long QT syndrome, underlying rare but life-threatening congenital heart condition.
However, the efficiency of the process leaves a lot to be desired even for such a good genome editor as CRISPR/Cas9. It is clear that CRISPR/Cas9 can be improved—after all, its first use for genome editing dates just 2013.
 The bottleneck of this process is in repairing the DNA break with the donor oligonucleotide as a template, because this reaction is rather inefficient. Fortunately, DNA repair is also among the major research interests of the Institute of Chemical Biology and Fundamental Medicine.
The bottleneck of this process is in repairing the DNA break with the donor oligonucleotide as a template, because this reaction is rather inefficient. Fortunately, DNA repair is also among the major research interests of the Institute of Chemical Biology and Fundamental Medicine.
One of the departments of the institute—the Group of Biopolymers Interactions—designs new constructs of donor oligonucleotides aiming to drastically increase the efficiency of genome editing. Moreover, DNA repair is also required in the conversion of differentiated cells into pluripotent ones; thus, this lab is involved in the work (supported by a grant of the Russian Science Foundation) on enhancing the iPS cell generation as compared to the classical Yamanaka procedure.
Imagine now that we have succeeded in correcting the cells and transplanting them back into the patient, where they start to interact with the whole body. It is small wonder that RNA is mixed up here as well. It has been discovered very recently that cells release exosomes, particles composed of protein and RNA, into the blood and other biological fluids. It looks like these particles are part of an absolutely new intercellular signaling mechanism, when a cell sends part of its genetic material to other cells of the body. However, neither the mechanism of exosome assembly nor the principles underlying protein and RNA selection for these particles are known yet. All these issues are also in the focus of attention of Novosibirsk researchers.
It would not be correct to state that RNA is the most important molecule of life. There would be no life either without DNA or proteins, and the “RNA world” must have come to an end long ago had Mother Nature not devised the currently existing split of the functions between DNA, RNA, and protein.
However, it goes without saying that over the last quarter century RNA turned from a poor cousin into an outstanding member of this triad. The research carried out at the Institute of Chemical Biology and Fundamental Medicine is intended to make the most of the numerous functions of RNA in targeted action on cells, which will enable developing new highly efficient medical technologies.
References
Vlassov, V.V. and Vorob’ev, P.E., RNA world: yesterday and now, Science First Hand, 2015, no. 1 (40), pp. 6–15.
Gorman, C and Fine Maron, D., The RNA revolution, Scientific American, 2014, vol. 310, no. 4, pp. 52–59.
Grigirivich, S., Was RNA in the beginning? In search for the initial molecule of life, Nauka i Zhizn, 2004, no. 2, pp. 44–55.
This work was supported by the Russian Science Foundation, project no. 14-24-00093