GMO and other genetic secrets of plant selection
For many years, plant cultivation worldwide has been enjoying the benefits of gene engineering – fast-growing, productive plants resistant to pests and extreme temperatures. However, to the average consumer, the letters “GMO” on the package are like the mark of Cain. Why is that so? The answer lies on the surface: gene engineering does what nature could never do – at least in the foreseeable future, – which is both impressive and scary. People are concerned that even the creators of GMOs themselves are reluctant to dismiss the risk of possible negative consequences of their cultivation and use. On the other hand, recent surveys by the Levada Center * have shown that only 30% of Russians know that all plants, and not only the genetically modified ones, contain genes, and that the rampant GMO-phobia is caused mainly by the total “genetic” illiteracy. Meanwhile, the range of modern methods of selection used to create new plant cultivars is very broad, and there are approaches that are no less risky than gene engineering, as well as others that are virtually unknown to the public
GMO stands for genetically modified organism, implying there has been some sort of influence on the plant genome – the storage of hereditary information, the sancta sanctorum of living cells. Today, the majority of the newest methods of plant selection gravitate towards direct DNA editing. By the definition provided by the WHO, this results in GM-plants – new varieties that could not have appeared naturally, i. e. as the result of breeding or natural recombination (“gene jumbling”).
While all of this is true, there is nothing revolutionary in modifying plant genomes! Humans have always influenced plant genetics, one way or another, without having the slightest idea about genes and their functions.
The path to modern cultivars that we eat began about 10 thousand years ago, with the emergence of agriculture. Humans selected the most robust and edible plants and grew them in an organized manner. In agriculture, there is no place for natural selection: according to artificial selection, only those plants survive that suit the humans’ needs.

Later, scientists discovered ways to influence DNA directly: in the 1960’s – by provoking mutations, and in 1983 – by using artificially created DNA. Nevertheless, traditional selection methods, such as crossing, may have changed significantly, but are still used on a par with the newest methods. It is the variability of approaches that gives the modern selection experts the most efficient set of tools to created “tailored” plants of the future
A telling example of a deviation between the purposes of natural and artificial selection is maize. In wild ancestors of modern corn cultivars, seeds separated from the cob easily, and fell to the ground. Such maize reproduced effortlessly, but the peasant always lost the bulk of the crop. What do we see now? In modern corn, seeds are attached to the cob securely even when it ripens. It is the same with other grains – rice, barley, or wheat.
 These cultivars are essentially the result of genome modification using various methods, such as crossing different varieties, which produces completely new cultivars. Mutations are a major resource for both artificial and natural selection: spontaneous mutations (changes) in plant DNA occur on a regular basis, caused by solar radiation and other factors. If such a mutation results in individuals with noticeable beneficial traits, they need to be reproduced – this sums up the selection process. The great variety of vegetables of the family Brassicaceae are a good example: broccoli, cauliflower, Brussels sprouts all share the same wild ancestor (Kempin et al., 1995).
These cultivars are essentially the result of genome modification using various methods, such as crossing different varieties, which produces completely new cultivars. Mutations are a major resource for both artificial and natural selection: spontaneous mutations (changes) in plant DNA occur on a regular basis, caused by solar radiation and other factors. If such a mutation results in individuals with noticeable beneficial traits, they need to be reproduced – this sums up the selection process. The great variety of vegetables of the family Brassicaceae are a good example: broccoli, cauliflower, Brussels sprouts all share the same wild ancestor (Kempin et al., 1995).
And there is more to selection. In the past 80 years, humans created over 3 thousand new cultivar varieties by treating the original forms with radiation and chemicals to cause unpredictable mutations in their DNA. Plants obtained as the result of such artificial undirected mutagenesis are still successively cultivated today. Moreover, counterintuitively, they have never been considered GMO. Eventually, the public has been lead to believe, incorrectly, that the first genetically modified plants appeared in the result of using gene engineering methods of precise DNA manipulation.
In any case, a new variety should be evaluated based on its properties, and not on the method of its creation. To have an opinion on the dangers of GMO, one must have at least a basic understanding of their origins.
A recipe for GMO: cut, fix, and stitch together
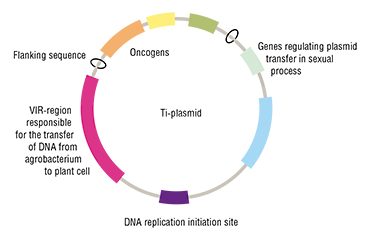
How do you make a GMO from a regular plant? The recipe is relatively easy. Take a plant cell genome and add a “genetic construction” – a DNA sequence coding the production of the necessary protein. You can add genes using a vector – a DNA or RNA molecule that can “proliferate” and transfer foreign genetic material from cell to cell, for instance, using a vector based on the circular bacterial plasmid.
This may sound simple, until you start thinking about how you insert the new genetic fragment precisely into the required region of the plant DNA cell. This is the most difficult problem of genome editing aimed at producing modern GMO’s.
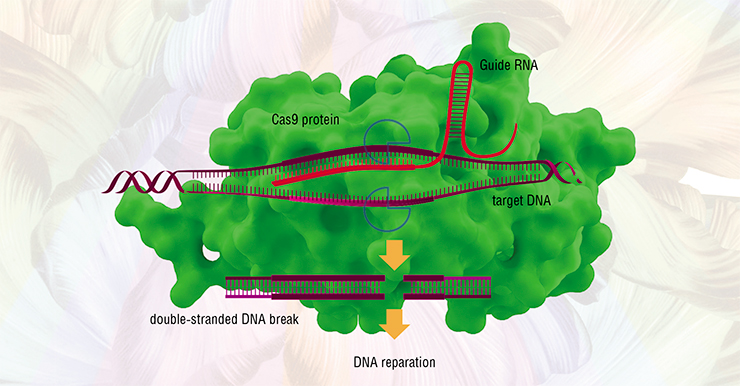
To split DNA molecules precisely in the necessary regions, scientists used restrictases, or “scissor” enzymes, capable of recognizing specific sequences of nucleotides – the building blocks of DNA. To stitch the DNA back together, other enzymes, called DNA-ligases, were used – their purpose is to repair damages in the DNA structure.
Just like 30 or 40 years ago, these methods are actively used today to obtain new variants of bacterial and viral genomes. However, these tools proved insufficient for working with genomes of higher organisms, such as plants and animals, including us humans. The problem lies in the ability of restrictases to recognize short DNA sequences, which is enough for efficient splitting of short bacterial DNA chains, where such regions are rare; however, in higher organisms, genomes contain multiple copies of short DNA sequences recognized by restrictases, which greatly reduces the accuracy of this method.
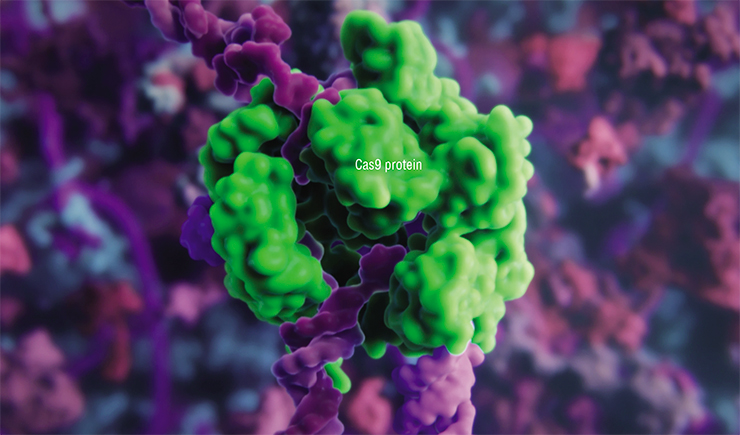
To edit such genomes, researchers had to create specialized tools for pinpoint DNA editing: first, it was oligonucleotide-directed mutagenesis of plants, followed by site-directed mutagenesis using nuclease enzymes with “zinc fingers”, TALENSs-nucleases and even meganucleases (Zakiyan, 2014; Daboussi, 2015). However, it was only when the famous CRISPR/Cas9 technology was discovered in 2012–2013 that scientists got close to pinpoint correction and editing of genes and genomes (Cong, 2013). The possibility of making controlled changes in the genetic information of living cells became a massive breakthrough, causing a number of global changes in selection.
How sharp are the genetic scissors?
The CRISPR/Cas9 system is based on a peculiar molecular mechanism that bacteria use to defend against bacteriophages (viruses affecting bacteria). When a pathogenic virus invades a bacterium, it triggers an “immune” reaction causing the foreign genetic sequence to split. This is done by Cas, the “scissors” protein, once the invader has been identified using its genetic “portrait” – fragments of viral DNA stored in the CRISPR region of the bacterial genome.
Based on bacterial CRISPR/Cas systems, scientists have created simplified artificial molecular structures that contain the Cas9 protein and offer exceptional precision in splitting DNA strands (Zakiyan, 2014). This has made it possible to perform all imaginable genomic modifications: introducing point mutations, inserting, correcting or removing long DNA sequences and fragments of selected genes.
The CRISPR/Cas9 system has already made it possible to make precise modifications in a variety of plant genomes; the results include new varieties of rice resistant to Xanthomonas bacterial rot as well as the famous “golden rice”, which carries the gene for beta-carotene (Chen, Gao, 2013). Another challenge has also been solved – creating plant-based “biofactories” capable of synthesizing human proteins: insulin, which is indispensable for diabetic patients, and albumin, used in the treatment of burns and cirrhosis.

Despite the proven efficiency of the CRISPR/Cas9 system, there remains a risk of nonspecific effect on the DNA and disruption of coding gene sequences. The explosive effect of the work published by a Chinese team of researchers from the Sun Yat-sen University, who used CRISPR/Cas9 to fix the genomes of human embryos to cure thalassemia, a hereditary disease, was hardly surprising, even with the declared success rate of only four out of 86 fertilized eggs (Liang et al., 2015).
Technologies that use the principles of synthetic biology are still facing substantial barriers of social cont-roversy, despite the high level of safety demonstrated by modern GM products. A few years ago, a massive outcry was spurred by reasonably doubtful research on the development of cancer in rats and death of butterflies provoked by the use of GMO (Walker, 2006). A survey conducted by Hart Research Associates in the USA in 2010 demonstrated that less than 10 % of respondents connect gene engineering with the threat of bio-terrorism or harm to the environment or human health; neither they believe that artificial creation of life is amoral. At the same time, a public opinion survey in the UK showed that over a half of the population understands the benefits of new biotechnologies to the society, despite the related ecological risk (Philp et al., 2014). The level of biological literacy in Russia can be illustrated by the results of a survey conducted in 2015 in the city of Kazan, where 55 % of the respondents supported the full ban of GMO, and only 15 % admitted that they are not exactly sure what GMO is, and the majority (48 %!) suggested that “all foods containing genes be banned from the market” (Business Online, 2015). Considering that genes are present in the cells of all living organisms, including bacteria, these vocal advocates of “genetic purity” are facing a bleak gastronomic future: an unchanging meal of buttered starch and sugared water, since even milk contains somatic cells with their genetic contentsToday, a number of experts are calling for a moratorium on all experiments connected with editing the genes of human embryos and gametes. Their concern is understandable: when it comes to the human genome, success must be guaranteed. However, progress is unstoppable: recently, Great Britain became the first state to allow this kind of experiments (Ershov, 2016).
Nevertheless, there has not been any decline in the human fear of invading the genomes of live organisms; in some cases, it keeps growing. This is the reason why there is strict legislation controlling the production, sale use of products of plant genome editing, which is keeping the global agriculture from transferring to advanced methods of selection. Nevertheless, scientists are not giving up: there are options that can minimize or completely exclude possible risks of negative consequences of introduction novel genes into plant organisms.
Lowering the risks: from TRANS to CIS and below
Today, all tests of the biological safety and marketing of genetically modified organisms, including plants, are subject to strict international regulations. The EU legislation regarding this issue is based on the Directive 2001/18/EC of the European Parliament and of the Council of 12 March 2001 on the deliberate release into the environment of genetically modified organisms. Notably, this document excludes organisms created by breeding, in vitro fertilization, polyploid induction, spontaneous mutations and merging protoplasts of two species (somatic hybridization) from the list of GMO organisms.
 In the Russian legislation concerning GMO, there are 4 federal laws and 6 government decrees, including the federal law #86-FZ “On state regulation in the area of gene engineering activities” dated July 5, 1996. Another governmental decree legalizing the cultivation of GM cultivars in Russia is due to come into effect; currently, such cultivars are only allowed for cultivation in experimental plots. 22 lines of food and forage GM plants are currently allowed for import, including corn, potatoes, soy, sugar beets and rice; all GMO and GM products are subject to obligatory registration.
In the Russian legislation concerning GMO, there are 4 federal laws and 6 government decrees, including the federal law #86-FZ “On state regulation in the area of gene engineering activities” dated July 5, 1996. Another governmental decree legalizing the cultivation of GM cultivars in Russia is due to come into effect; currently, such cultivars are only allowed for cultivation in experimental plots. 22 lines of food and forage GM plants are currently allowed for import, including corn, potatoes, soy, sugar beets and rice; all GMO and GM products are subject to obligatory registration.
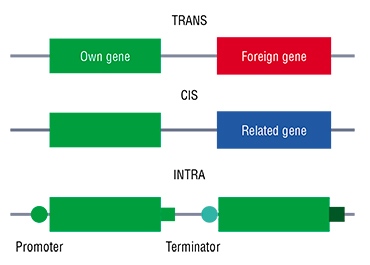
The global scientific community in its turn believes that various GMO’s must be distinguished by their origin, and that products acquired using moderate invasion into the genome must be less regulated. This gave rise to the system of separating GMO’s into three types: TRANS, CIS and INTRA.
 Trangenic organisms have artificially inserted genes that cannot be acquired by natural breeding. This includes genes from other plants or animals, e. g. rice with corn genes. The potential danger of transgenic cultures lies in the possibility of the acquired traits affecting the use of the plant as human food or livestock forage, or in the transfer of these traits to wild relatives with unpredictable consequences to the ecosystem. This is the reason why in developed countries, legislation focuses on the biological safety of such cultivars to lower the risk of ecological shifts.
Trangenic organisms have artificially inserted genes that cannot be acquired by natural breeding. This includes genes from other plants or animals, e. g. rice with corn genes. The potential danger of transgenic cultures lies in the possibility of the acquired traits affecting the use of the plant as human food or livestock forage, or in the transfer of these traits to wild relatives with unpredictable consequences to the ecosystem. This is the reason why in developed countries, legislation focuses on the biological safety of such cultivars to lower the risk of ecological shifts.
With cisgenic plants, genes from the same species or closely related species with which the plant can produce offspring in the wild are inserted into the genome. The target gene must not be modified or separated from its regulatory sequences. Potatoes resistant to rot thanks to the insertion of genes from wild Andean potatoes resistant to that disease are a good example of a cisgenic plant. Belgian researchers are working on such a variety now (VIB’s fact series, 2015). It is important to note that cisgenesis does not add any new traits to the plant organism and is essentially analogous to the traditional crossing of cultivars with their related wild forms.
Intragenesis can be considered a development of the cisgenesis concept, however, in this case, the plant DNA is inserted with its own gene, combined with regulatory regions from its other genes. Modifications of this sort artificially create new combinations of existing DNA regions (Holme, 2013). Such modifications of gene activity regulations allows to enhance useful traits (such as the ability of a plant to accumulate vitamins in its leaves), or, conversely, to remove or minimize undesired traits.
Meanwhile, modern GMO regulations do not take into account the differences between transgenic and cisgenic plants, even though they are radically different. Strict legal boundaries complicate the production and use of cisgenic plants to the extent of blocking or significantly delaying future research on the improvement of agricultural cultivars. Only in Canada, cisgenic plant control is less severe compared to transgenic plants (Schouten, 2006).
Somatic Frankenstein
Curiously, the mighty legislative shield against GMO has holes due to a number of paradoxes and allowances that favor daring selectioneers. Somatic hybridization is a good example. It deals with the formation of new plant forms by combining nuclear and other (mitochondrial and plastid) genes during the cultivation and merging of common somatic cells that make up plant tissues and do not take part in sexual reproduction. This type of plant hybridization is relatively widespread; in the EU, such somatic hybrids are not considered GMO, meaning their production and sales are not subject to strict regulation.
What is this magic method of selection? At the first stage, cells of different plant species (as a rule, a cultivar and a wild plant) are treated with special agents that destroy the cell wall and obtain the protoplasts. Next, protoplasts are forced to stick together or merge using chemical or mechanical methods, causing them to restore a shared cell wall. As the result, two or more parental cells give rise to a new organism, called a regenerant, or somatic hybrid.
The fate of the parental genes in this case can be different. Two nuclei can divide simultaneously without merging, forming binuclear daughter cells. If they merge during mitosis, stable mononuclear daughter cells are formed, carrying mixed genetic material. As for the extranuclear genome, it can come from one parent or be mixed. Somatic hybridization allows the creation of all sorts of hybrids, including hybrids that could never have appeared sexually, such as hybrids carrying cytoplasmic genes from both parents, and not only from the mother; or “cybrids”, which get their nucleus from one parent and their cytoplasm – from the other, etc.

Using somatic cells in hybridization allows scientists to work with genetically distant species that cannot be bred normally, or with completely sterile plants. In other words, this method is used when there is a need to overcome an incompatibility between cultivars and wild forms. This way, it is possible to create interfamilial, interclass, or even interkingdom hybrid colonies, such as rice + soy, barley + tobacco, and even tobacco + mouse (Makonkawkeyoon, 1995)! To be fair, the majority of such regenerants are incapable of reproduction and often exist as cell clusters rather than a true organism.
Curiously, while somatic hybridization causes a substantial gene jumbling, and its results are greatly unpredictable, it is still allowed for use in agriculture, unlike the methods of directed mutagenesis. “Curiouser and curiouser”, as Alice said…
What hides behind a graft
Let us have a look at methods that will satisfy even the most stern adepts of natural foods. These methods have been in use for a long time, and have not encountered any social or legal resistance. It turns out that from the point of view of genetics, they are not entirely “without sin”, and combining them with cutting-edge approaches opens unsuspected perspectives.
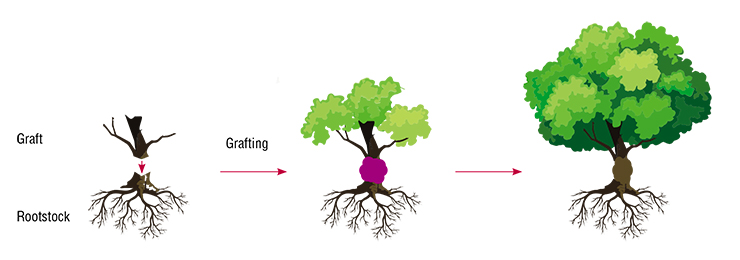
For instance, grafting is one of the long-known and ubiquitous methods of plant reproduction encountered by gardening enthusiasts. It consist in grafting a stem from one plant (called graft) onto the root system of another plant (called rootstock). At the end of the XIX century, this method helped to save European varieties of grapes (Vitis vinifera) from a massive infestation of Phylloxera, an insect pest that damages roots. Vines were grafted onto rootstocks of a North American grape species, Vitis labrusca, resistant to this pest (Troshin, 1999). In 2003, a R. Baur, a farmer from Oregon, created an actual “tomacco” (tomato + tobacco), like Homer from “the Simpsons”. Tests showed it to contain nicotine, albeit in the leaves and not in the fruit (Philipkoski, 2003).
What happens with the plant when it is grafted, if the genomes of both the graft and rootstock remain unchanged? First, the size of fruit, growth speed, and ripening time can changee in the rootstock. These new traits are not passed on to the progeny through seeds, because they are not hereditary. Second, grafting can result in chimera changes, and the grafted plant will consist of genetically different plants. This is often used with ornamental plants to create plants with variegated coloring of their leaves or petals.
Grafting can cause real mutations, provoked by certain chemicals (ethylmethanesulfonate, ethylimine, etc.), which move from the rootstock to the graft. However, mutations after grafting are very rare. An undoubted benefit of grafts is the possibility to reproduce mutations that cannot be inherited, while the main disadvantage lies in the large volume of the necessary source material.
Grafting is indeed a tried and safe method. But what happens if we use a GM plant for rootstock? Will the resulting plant be a GMO? The law says not: fruit from such hybrids are not listed as GMO, because the DNA of the graft remains unchanged. However, we cannot be sure that there is no exchange of genetic information between the rootstock and the graft. for instance, RNA molecules regulating the work of the genome can be passed up from the rootstock, meaning that the levels of production of certain proteins in the graft cannot be predicted.
Giving the genes a boost!
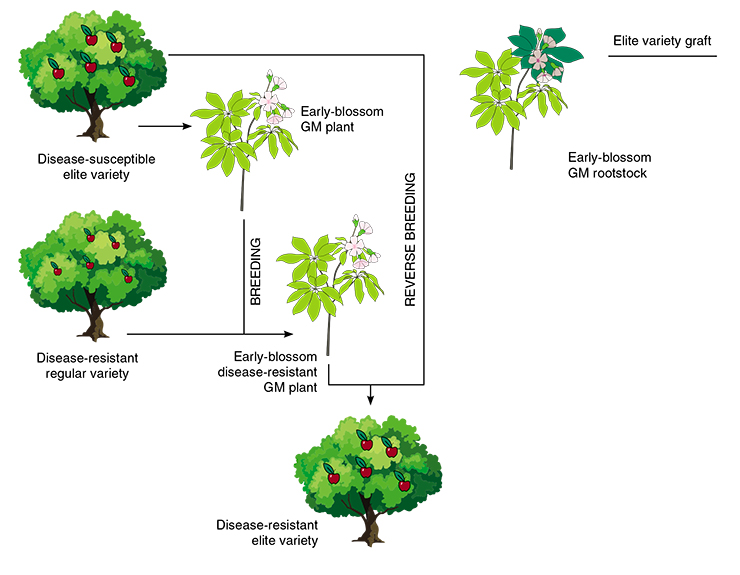
Grafting is not the only roundabout way to create new varieties with modified DNA activity. Fast-track breeding of trees and shrubs is a whole complex of methods and techniques aimed at speeding up the development of new varieties, which is especially important for perennial cultivars. The reproduction cycle of trees with large fruit, such as plums or hazelnut, can take up to a decade and more (van Nocker, 2014). This means that the breeder has to wait 5 to 10 years after planting a hybrid seedling until it matures to be able to continue the work. If there is a need for several consecutive crossings, creating a new variety may take up to 30 years, and no one is ready to wait for so long in the modern world.

To speed up this process to the maximum, scientists have been spraying their test subjects with growth hormones, growing them at high temperatures, and using other tricks, including DNA technologies. One of the safest methods is the so-called marker-assisted selection, which consists in analyzing the genomes of new sprouts or even seeds and selecting the best hybrids long before they turn into mature plants. Today, there is no need to test the plant with pathogens to check their resistance – one can simply detect the necessary gene in the seed. The main flaw of this method is the high cost, because DNA screening is not cheap.
 Breeders use a variety of tricks to speed up the maturing of their plants. In some cases, they artificially activate genes responsible for setting off the reproduction process, causing a young plant to bloom and fruit. Sometimes, additional genes are added to tree genomes to speed up blooming and fruiting, and seedlings begin to bloom after just one year. Combining the methods of accelerated and reverse (when a hybrid is bred to one of its parents) breeding the fast blooming gene can be introduced into the original variety and remove it at the last step of the selection process by breeding the genetically modified hybrid to the parent plant.
Breeders use a variety of tricks to speed up the maturing of their plants. In some cases, they artificially activate genes responsible for setting off the reproduction process, causing a young plant to bloom and fruit. Sometimes, additional genes are added to tree genomes to speed up blooming and fruiting, and seedlings begin to bloom after just one year. Combining the methods of accelerated and reverse (when a hybrid is bred to one of its parents) breeding the fast blooming gene can be introduced into the original variety and remove it at the last step of the selection process by breeding the genetically modified hybrid to the parent plant.
Fast-track breeding is also done by grafting to a GM rootstock. In this case, the secret lies in the active work of genes responsible for blooming in the genetically modified rootstock. As the result, the graft part receives specialized proteins that trigger the maturation mechanism, and blossoming begins.
Modern methods of grafting and fast-track breeding are hiding a wealth of genetic secrets behind their plain and traditional appearance. At the same time, scientists facing the public controversy and strict legislative regulations around the creation and use of GMO’s are trying to avoid making direct changes in plant DNA. This brings us to the most enigmatic group of modern methods of selection.
Epigenetics: a little bit does not count
One of the newest and cutting-edge alternatives to selection consists in using epigenetic approaches. Epigenetics studies inherited mechanisms of gene expression control (Marjori, 2015). How our genetic code works is common knowledge, however, the peculiarities of its “fine tuning” (epi- means “above”), which acts as a music conductor, for the most part remain a mystery.
The beginning of protein production in the cell is regulated by a multitude of factors. The cell also has ways to “silence” a specific gene to prevent the production of a protein that is no longer necessary: in includes destroying unripe RNA molecules that have been read from the genetic “matrix”, or creating mechanical obstacles for the very process of DNA reading (Marjori, 2014). Overall, epigenetic signals in the cell are numerous and poorly understood, however, yet some of them are already used for the selection of plants that we see on our plates every day.

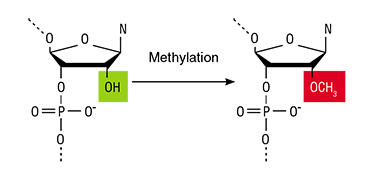
One of the ways to suppress the work of genes in the cell is by using a natural mechanism called RNA-dependent DNA methylation, which consists in the binding of a methyl group (СН3) to the nucleotide cytosine in certain positions. As a result, the transcription of DNA to RNA is blocked (Zhang, 2013).
DNA methylation in plants and animals is done by enzymes called DNA-methyltransferases. These enzymes themselves cannot methylate DNA: they need special noncoding RNAs that direct the methyltransfelases to specific DNA regions. Moreover, there are two more RNA types believed to be involved in DNA methylation: small interfering RNA and microRNA. Together, these molecules define which region of the DNA chain will be methylated. Today, such RNA can be injected intothe plant using a variety of techniques, such as plant viruses or by means of gene engineering techniques (Deng, 2014).
Interestingly, if a scientist changes a plant trait using DNA methylation without introducing any mutations to the genome, the resulting plant is not considered a GMO. If the noncoding RNA are not injected from the outside and are produced by the plant itself using genomic editing, such plant is considered to be genetically modified.

There is a way to trick our way around this problem. In plants, methylation of some regions of DNA can be inherited, i. e. transmitted vertically from parents to future generations (Jones, 2001). By breeding a GM plant with its natural form it is possible to obtain a hybrid that has no modified DNA, but retains the methylation. Such hybrid will not be considered genetically modified.
How safe is methylation? Quite safe, since places where methyl groups bind to DNA are not completely random. Unlike traditional selection methods, this type of intervention has predictable results: we can select a gene coding a specific protein and simply silence it. However, this must be done carefully, since DNA methylation mechanisms are rather complicated. Otherwise, this can result in a plant prone to diseases or premature aging.
In some cases, DNA methylation is actually required for a gene to begin working. Scientists have harnessed this process: using DNA methylation, it is possible to increase the activity of genes responsible for the production of storage proteins. For instance, methylation makes it possible to increase protein content in wheat grain, and treating rice with a methylation inhibitor (5-azaticidin) produces plants with inheritable dwarfism (Vanyushin, 2013).
RNA silence as an allergy shield
A successful reading of a gene to matrix RNA does not mean that the protein that it codes will be built: this mRNA can end up being destroyed in the cytoplasm. This phenomenon, called post-transcriptional silencing, is often observed when additional genes are inserted into plant DNA. It was described back in 1990, when additional copies of a gene responsible for the red color of blossoms were inserted into the petunia genome, and, contrary to the expectations, the amount of the red pigment decreased substantially (Napoli et al., 1990).
The mechanism of RNA silencing lowers the efficiency of gene engineering. On the other hand, it can be used to create plants resistant to plant viruses, because it can be helpful in the destruction not only of the plant’s own mRNA, but also of the corresponding RNA of viruses that have managed to invade plant cells.

Apparently, there exist several mechanisms of post-transcriptional silencing, and as of now, scientists do not have the full picture of their function and connection with each other; there are yet many blanks to be filled (Plotnikov, 2007). One of the hypotheses suggests that individual mRNA molecules actively degrade when they reach a certain threshold of their numbers (Abler, 1996). Another theory is based on changes in genome regulation connected to DNA methylation, resulting in the production of some “abnormal” RNA molecules along with normal RNA molecules, triggering the decay of mRNA in the cytoplasm (Hoofvan, 1997).
RNA-interference is one of the best-described mechanisms of post-transcriptional silencing. This method is based on the ability of double-strand RNA molecules to efficiently suppress genes with similar structure. In recent years, RNA interference has been used in applied research aimed at creating knockout (i. e. containing “silent” genes) cells, tissues and organisms. If used wisely, the methods allows to “switch off” the production of any protein in the cell.
An example of successful application of this method is the creation of two varieties of the coffee plant with 30–50 % less caffeine in the fruit. A similar experiment was carried out with tobacco to lower the nicotine content levels in the plant (Ryabushkina, 2009).
Another possible application of this approach is allergen synthesis suppression. This is science fiction made real: geneticists from Institute of agriculture in Cordoba managed to nearly completely remove gliadin, a component of gluten, from wheat grain. The gliadin group of storage proteins is what causes an immune reaction to wheat in many people. However, to achieve this, the scientists still had to use the CRISPR/Cas9 gene editing system (Sanchez-Leon et al., 2017).

Of course, there is a long road ahead of scientists working in this field; and yet there is hope that soon, people will be able to enjoy a peanut butter sandwich without the risk of dying from anaphylactic shock! We must remark that such invasion of gene engineering into plant metabolism would be different from the traditional approach: no foreign genetic material is added into the genome, meaning that no foreign proteins will be synthesized. This means that RNA-interference can be considered a low ecological risk variety of genetic intervention. Moreover, even formally, such intervention would not bear the “GMO” stamp.
Summing up, what is the takeaway of our tour of the modern methods of selection? The chance to choose “natural” grains, vegetables and fruit is long lost. Progress is unstoppable, and this is true about creating new plant species as well; however, one must understand and correctly gauge the risks of distributing and using genetically modified products.
Today, as the public is showing increased concern with food safety, selection experts are forced to use alternative methods to create new varieties. For various reasons the use of methods is not legally restricted, however, in some cases they are as risky as the methods of creating GMO’s.
One should keep in mind that a wise approach to plant selection using genome editing allows minimizing the use of pesticides and fertilizers – the ecological significance of that does not require any further explanation. In any case, what products will make up our diet tomorrow depends on us rather than on nature.
* key non-government public opinion monitoring organization in Russia
References
Cong L., Ran F. A., Cox D. et al. Multiplex Genome Engineering Using CRISPR/Cas Systems // Science. 2013. V. 339. P. 819–823.
From plant to crop: the past, present and future of plant breeding. 2016. VIB’s fact series, 44 p.
Matzke M. A., Kanno T., Matzke A. J. M. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants // Annu. Rev. Plant Biol. 2015. V. 66. P. 243–267.
Moghaddassi S., Eyestone W., Bishop C. E. TALEN-Mediated Modification of the Bovine Genome for Large-Scale Production of Human Serum Albumin. 2014, PLoS ONE. 9, e89631.
Philp J. C., Ritchie R. J., Allan J. E. Synthetic biology, the bioeconomy, and a societal quandary // Trends in Biotechnology. 2013. V. 31. P. 269–272.
Shumnyi V. K. Nature was the first gene engineer // Science First Hand. 2004. V. 2. N. 3. P. 32–39. (in Russian)
Zakiyan S. M., Vlasov V. V., Medvedev S. P. “Genome editors”: from “zinc fingers” to CRISPR // Science First Hand. 2014. V. 56. N. 2. P. 44–53. (in Russian)
This publication is based on a paper presented at the “bio/mol/text”-2017 popular science contest on the “Biomolecula” website (biomolecula.ru)