Influenza Virus Ion Shield
Perhaps it is hardly possible to find a person who has not caught flu at least once in lifetime. But most of us willy-nilly “deal with” this infectious agent over and over again. A great variety of medicines supposed to alleviate this disease, which seems harmless only at first sight, is another proof to that. Alas! Influenza is one of the diseases that are overcome in a week if they are treated and in 7 days if they are not.
But what makes influenza viruses so exceptionally resistant to drugs and our immune system attacks? The answer might lie on the surface in the literal sense of the word — on the influenza virion surface…
The influenza virus genome is known to be able to mutate at an exceedingly high rate because of accidental “errors” in the replication of its genetic information. Nucleotide substitutes are hundreds of thousands times more frequent in its hereditary material than in the human genome.
It is this virus peculiarity that serves as a basis for an effective mechanism protecting it from the human immune system, since it leads to the appearance of such strains that antibodies previously generated in a body in reaction to the infection or vaccination cannot bind with. Thus, a mutant strain actively propagates itself until the body generates new specific antibodies.
The molecular mechanisms that make it possible for the virus to penetrate into the target cell are still studied by many scientists. The main purpose of this research is to assist with the creation of medicines which could prevent a cell from the virus infection. It is imperative to stop the disease at the initial stage, that is, to prevent a virus particle from generating an army of its clones ready to contaminate other healthy cells.
Nowadays research into the type A influenza virus (most flu strains of mammals and birds are of this type) become even more important. This is due to a possible influenza pandemic which may be provoked by a mutated strain of the so-called bird flu. A profound understanding of how the complicated mechanisms of intermolecular interactions work and how both the virus and host genes activity is regulated is needed to create new effective means for the prevention and treatment of this disease.
Methods of computer modeling are an unusual and little-known approach in this field not only for the general public but also for many experts. They enable one to reconstruct the regularities of molecular evolution and run a kind of virtual “experiments” both with separate molecules and molecular-genetic systems. Owing to these methods it is possible to reveal molecular targets having great potential for the creation of a new generation of medical products against various viral and bacterial infections.
Computer methods and approaches in evolutionary biology and proteomics (protein science) have been actively developed at the Laboratory of Theoretical Genetics at the Institute of Cytology and Genetics of the Siberian Branch of RAS during the last 15 years. Lately one of the most important objects of research has become the influenza A virus or, more precisely, hemagglutinin — one of its surface proteins.
Hemagglutinin — the virus “grapnel”
Before we pass on to the results of computer modeling themselves, it is necessary to have a closer look at the structure and “mode of living” of the influenza virion, and especially at its structural and functional protein complexes.
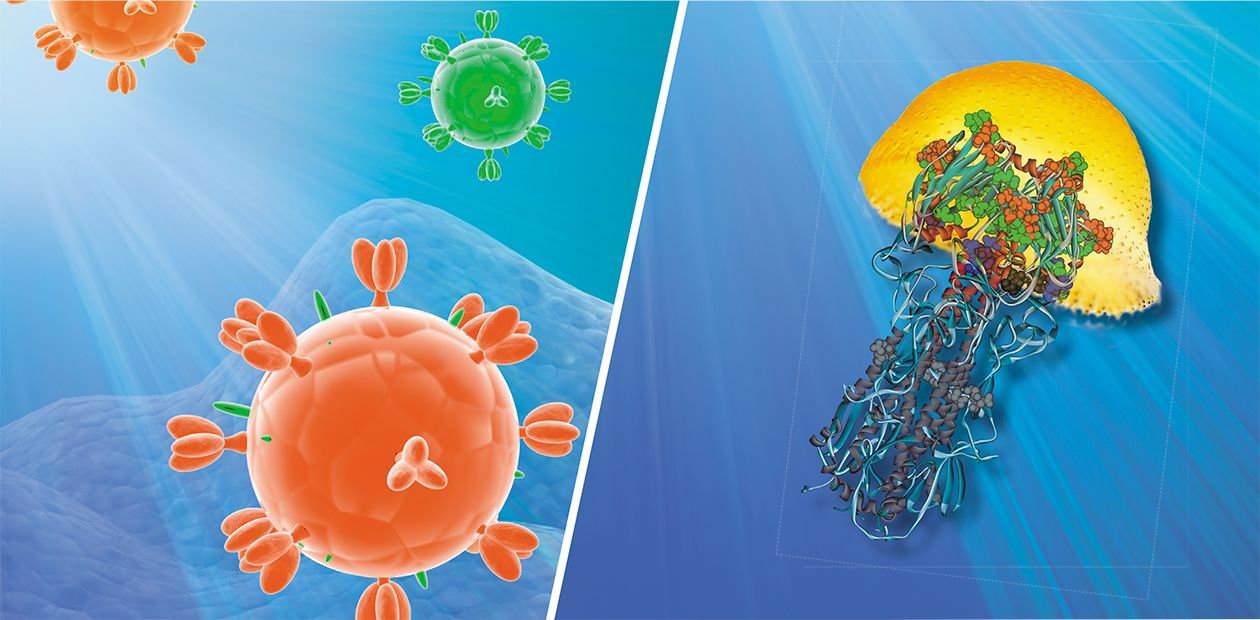
The virus genetic material is protected by a shell which consists of a special protein. It is encircled with a membrane made of lipids (lipoids) that keeps molecules of the following three proteins: hemagglutinin, neuraminidase, and M2 protein, the latter forming special ion channels. Hemagglutinin helps the virus to attach to the cell membrane while neuraminidase destroys it so that the virus can penetrate into the cell. Then the medium acidity (pH) inside the invader virion changes owing to the ion channels, which facilitates the liberation of genetic material from the virus capsule.
Thus, hemagglutinin plays an important role in the contamination of the target cell by the virus. It takes the form of trimers, i.e. three-molecule complexes, on the surface of the viral shell. Each molecule consists of two subunits. One of them mediates the viral-cell attachment; and the other is responsible for the cell membrane fusion. The process of fusion is also far from being simple. It is accompanied by considerable conformational changes of the hemagglutinin molecule, which result in the liberation of a special fusion peptide “hidden” in the inner part of the protein globule.
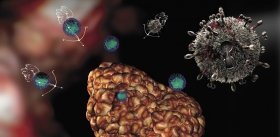
For the infected organism, surface virus proteins, including hemagglutinin, are of special importance, since they are antigens — substances that help the immune system to identify the infectious intruder. In the structure of the viral proteins one can single out so-called antigen determinants — the sites with which antibodies, specific defensive proteins of the immune system, preferably bind. It is mutations causing structural changes in such zones, i. e. changing the shape and location of the antigen determinants, that lead to the appearance of new virus strains. Such viruses become almost “invisible” for antibodies already circulating in the blood of the infected organism, which makes it defenseless against invasion.
Our supposition was that, besides structural mutations, there might be other mutations protecting viral proteins from the action of antibodies. The fact is that as result of mutations there appear new sites in the areas of antigen determinants. And ligands — molecules or ions that can form complex compounds with the protein — can bind with these sites. As a result, antibodies of the infected body cannot interact with such “well-protected” viral antigens. We have verified this hypothesis applying computer methods of molecular modeling developed in our laboratory.
What computer helped us to see
Basically, protein molecules can interact with other molecules of diverse nature: macromolecules, among which there may be proteins or nucleic acids, genetic information carriers; low-molecular compounds; and various ions. In this respect virus proteins do not differ from the proteins of other living organisms, including human beings.
Such universal interactions underlie metabolic pathways, transport molecular pathways, signal transduction pathways… There are special ligand binding sites — functional sites — on the protein surface. Their unique structure facilitates specific binding with these molecules.
And now let us come back to our “hero” — hemagglutinin — and look at the results of computer analysis. First of all, it should be noted that, in contrast to other proteins, hemagglutinin surface abounds in binding sites of various ions. Moreover, it has a great number of potential sites of this kind. And a single mutation, i. e. the substitution of only one nucleotide pair, can turn them into “active” binding sites!
Perhaps, it is these hemagglutinin properties that allow the influenza virus to easily escape our immune system and provide its high epidemicity. The fact that the identified ion binding sites almost overlap with antigen determinants sites is an argument in favor of this assumption.
The Laboratory of Theoretical Genetics at the Institute of Cytology and Genetics of the Siberian Branch of RAS has created a database (PDBSite) comprising information about 3D structures of more than 30 thousand different functional protein sites, including catalytically active sites, protein-protein, protein-DNA/RNA, protein-drug interaction sites, etc. Easy to search and operate with, the database stores data about the physicochemical, structural, and evolutional properties of these sites obtained by the computer analysis of experimentally resolved atomic structure of proteins. These data are successfully applied to recognize functional sites within the spatial structures of little-studied proteins by the tracing of their structural similarity with the already known proteins. For instance, after the structure of hemagglutinin — the influenza virus surface protein — was analyzed, it was found that one of its sites resembles the sulphate-ion binding site (SO4), which proves the potential ability of this protein to bind with the sulphate-ion at this site.
The antibodies generated as a result of the immune response to infection are known to bind with the antigen determinants. This leads to the neutralization of virus particles and their further destruction under the action of the immune system. Therefore, it is evident that the ion binding with an antigen determinant will impede the formation of a stable “antigen-antibody” complex. Consequently, the virus will be “shielded” from the influence of the immune system.
Our hypothesis of the influenza virus “ion shield” was confirmed by one more peculiarity of hemagglutinin that has been revealed: the assumed ion binding sites on its molecule coincide with the already known sites responsible for the adaptive molecular evolution of this protein.
Usually it is considered that adaptive mutations are those that increase the adaptive properties of the organism. Indeed, mutations causing damage to the antigen determinants are adaptive for the influenza virus. Apparently, the emergence of ion binding sites in these zones essential for the virus reproduction and survival plays an important role in the adaptive evolution of the influenza virus surface protein.
It was discovered that the spatial structure of the influenza virus surface protein contains numerous binding sites of various ions (as well as potential binding sites), which favors the formation of a certain ion shield around the virus protecting it from immune system attacks
The research results unambiguously testify that possible ion binding with the surface viral protein should be taken into consideration in the creation of a new generation of vaccines for the influenza virus neutralization. This very simple and effective virus defense mechanism, based on the results of single mutations, should receive proper attention. Other probable consequences of the “ion shield” formation should not be overlooked. For example, charged particles on the surface of the virus may facilitate its fusion with the cell membrane and penetration into the cell.
The above suppositions have made it possible to view from a new angle the problem of mechanisms of virus defense against the host’s immune system attacks and discover new molecules taking part in the penetration of the virus into a cell. These hypotheses surely need further analysis and experimental verification.
However, there is no doubt that ions may be important factors regulating the influenza virus life cycle. Nor is it questionable that the study of such molecular mechanisms will contribute to the discovery of new pharmacological targets and the creation of effective vaccines and medical products.