Bacteriophage, We See You!
A century has elapsed since Frederick William Twort, a British microbiologist, noticed transparent glass-like spots formed of dead bacterial cells in colonies of micrococci. For a long time after their discovery, the research into bacteriophages was of a phenomenological character because of the lack of adequate experimental tools. The scientists had no possibility for studying in detail the specific features of an antibacterial impact caused by bacteriophages since they are invisible not only to the naked eye, but also to light microscopy. The advent of electron microscopy brought the study of viruses, including the viruses of bacteria, to a fundamentally new level
Electron microscopy studies suggest that bacteriophages are actually nanoorganisms rather than microorganisms, since their size does not exceed 100 nm. Also, it has been shown that bacteriophages are most diverse in their structure. This raises the question of their nomenclature. The very first classification, dating back to 1943, was based on the specific structural features described with the help of electron microscopy. One of its founders, Ernst Ruska, distinguished three types of bacteriophages, using their morphological characteristics (Ackermann, 2009).
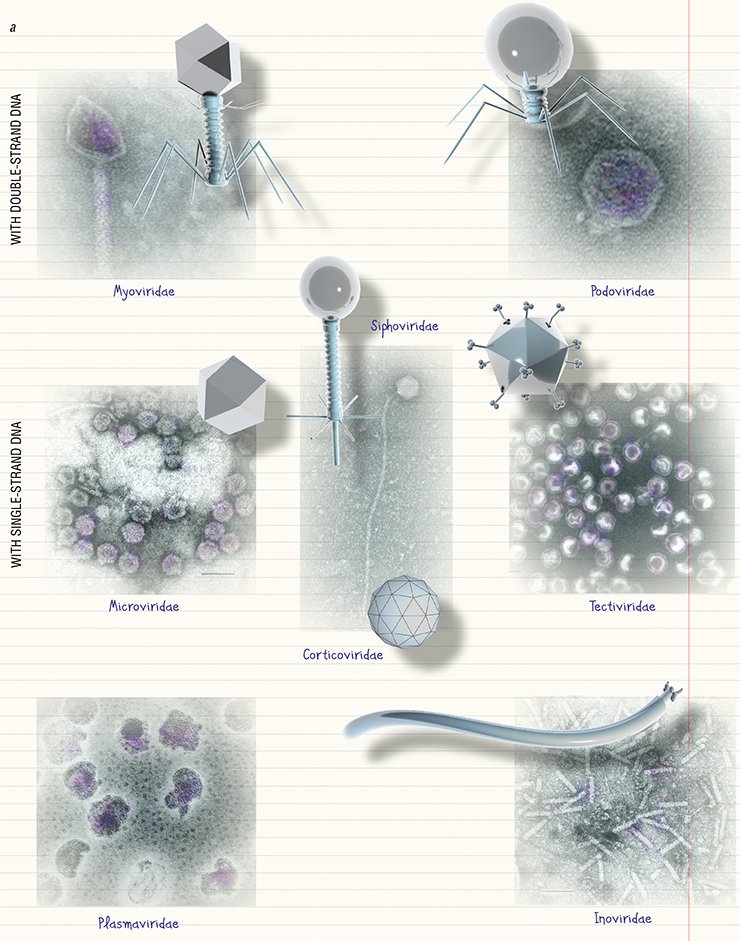
The currently used systematics of bacteriophages, developed in 1967, relied on the classification that comprised six morphotypes. However, the discovery of new bacteriophages has added new families, genera, and species to this systematics. The advances in molecular biological tools established additional criteria for their classification which took into account the type of nucleic acid and/or protein composition within the phage.
The use of state-of-the-art molecular techniques in the bacteriophage research has revealed manifold specific features of these interesting organisms. In turn, bacteriophages turned out to be a most useful methodological tool for molecular biologists (Brussow, 2013).
If there is a head, there is a tail
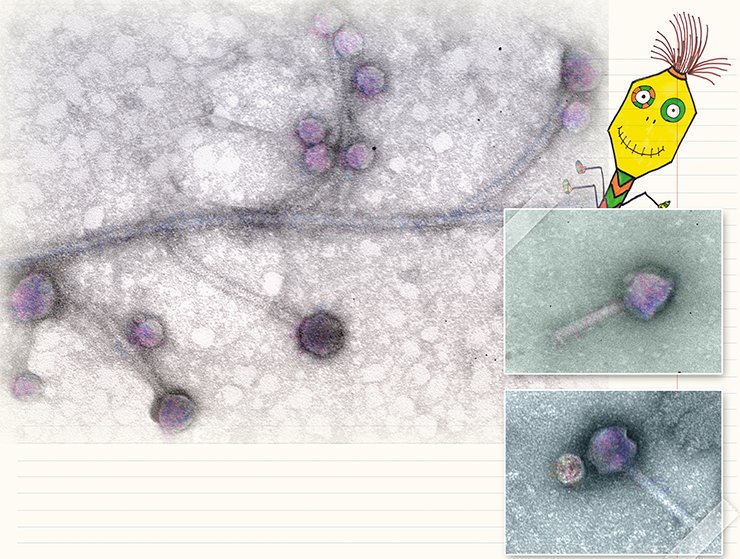
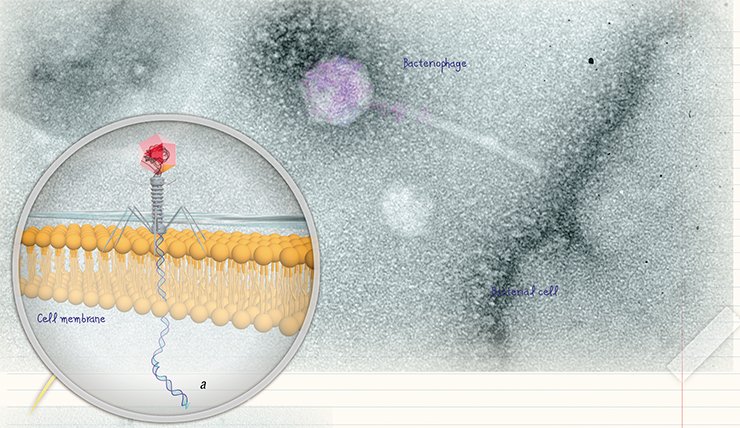
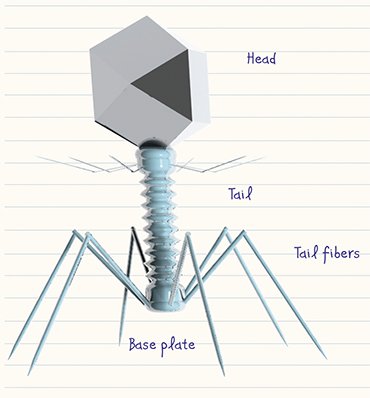 Actually, bacteriophages have a comparatively simple structure: each virus is a complex of a nucleic acid and proteins packaged in a specific manner. They may be curiously shaped but approximately 96 % of all known bacteriophages have a “tailed” phenotype (Matsuzaki et al., 2005). They have a “head” shaped as icosahedron (a protein reservoir housing packaged nucleic acid) and a “tail,” a protein structure carrying the elements that are able to bind stably to receptors (specific proteins or polysaccharides) on the surface of bacteria. Individual species of “tailed” bacteriophages differ in the size of their “head” as well as in the size and fine structure of their “tail.”
Actually, bacteriophages have a comparatively simple structure: each virus is a complex of a nucleic acid and proteins packaged in a specific manner. They may be curiously shaped but approximately 96 % of all known bacteriophages have a “tailed” phenotype (Matsuzaki et al., 2005). They have a “head” shaped as icosahedron (a protein reservoir housing packaged nucleic acid) and a “tail,” a protein structure carrying the elements that are able to bind stably to receptors (specific proteins or polysaccharides) on the surface of bacteria. Individual species of “tailed” bacteriophages differ in the size of their “head” as well as in the size and fine structure of their “tail.”
A bacteriophage species is identified according to its ultrastructural characteristics, which are described using the negative contrast technique. Any phage-containing suspension can be used as a sample, be it water from a natural source, animal intestinal lavage specimens, or bacterial cell suspension after incubation with a bacteriophage in a laboratory. A drop of prepared suspension is covered with a special copper grid with a polymer film on it for the bacteriophages to sorb. The grid is then treated with a counterstain (usually uranyl acetate or phosphotungstic acid) that encompasses bacteriophage particles and creates a dark background, rendering bacteriophages, which have low electron density, visible in the electron microscope.
Over 6300 bacteriophages have been described by electron microscopy so far (Ackerman and Tiekotter, 2012; Ackermann and Prangishvili, 2012). It emerges that not all bacteriophages have clearly distinguishable “head” and “tail”; as for their hereditary matter, the phages with double-strand DNA are the most abundant. The systematics of bacteriophages is very dynamic since novel phages are constantly being discovered (Ackermann, 2007).
Hunting for bacteria
Advances in electron microscopy techniques made it possible to visualize not only bacteriophages, but also their reproduction. The penetration of “tailed” bacteriophages into the cell has been studied in most detail, including the molecular mechanisms underlying the “injection” of phage DNA into the cytoplasm of a bacterial cell (Guerrero-Ferreira and Wright, 2013).
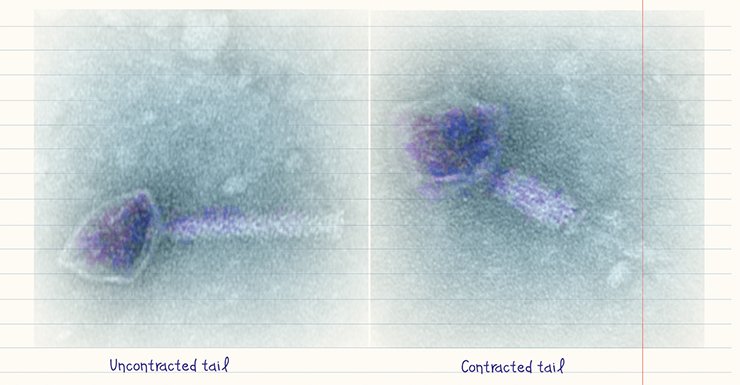
The typical behavior of bacteriophages “attacking” a bacterium is demonstrated by a lytic phage. The phage attaches itself to the bacterium surface, using its receptors as an “anchor.” Then its “tail” pierces the bacterial wall with the help of specialized proteins to form a “channel” for the phage nucleic acid to pass into the cell. Within half hour, the protein and nucleic acid components ofthe bacteriophage are synthesized in the infected bacterial cell to assemble new phage particles. Then the cell is destroyed to release mature virions.
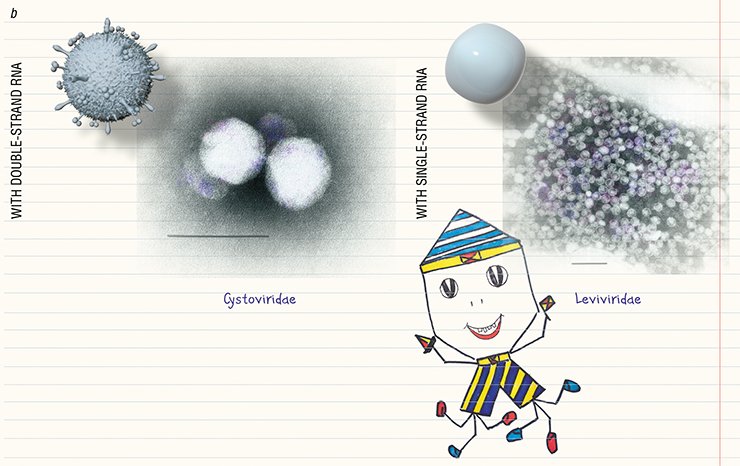
The unique characteristics of archaeophages along with the three cell lineages found on the planet—bacteria, archaea, and eukaryotes (the organisms possessing a cell nucleus)—suggest that there exist three specific virus “domains” that have emerged in the course of a long-term coevolution of viruses and their hosts, although some viruses still retain the traces of their common origin (Pina et al., 2011)
The combination of counterstaining and ultrathin sections* makes it possible to trace all stages in bacteriophage reproduction including the sorption of phage particles on the surface of bacterial cells, their penetration into the cell, and copying. Unfortunately, this approach is much less developed as compared with bacteriophage visualization and identification by counterstaining. And yet, the ultrastructural characteristics of the bacteriophage life cycle are helpful in assessing the efficiency of elaborated phage therapies satisfactorily.
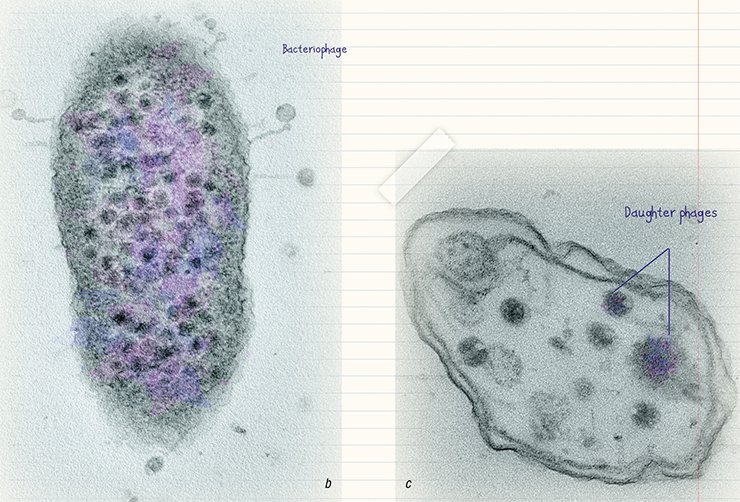
Bacteriophages are undoubtedly a unique phenomenon on the planet: on the one hand, they have a rather simple structure; on the other hand, they are tremendously diverse both in terms of their morphology and potential “victims.”
These nanoorganisms are not just safe for us – they are “friendly” because they can kill pathogenic bacterial cells without affecting any cells of higher organisms, including the cells of humans, agricultural animals, and plants. This property allows us to use bacteriophages for treating bacterial infections following the principle that the enemy of our enemy is our friend.
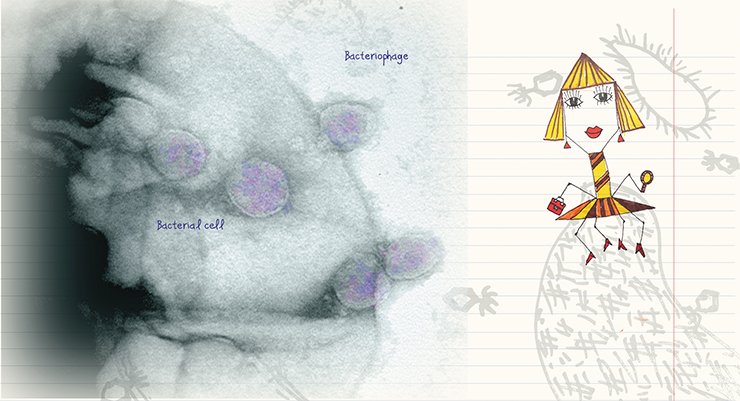
Phage therapy has agreat potential not only because phages can kill bacteria, but also because of a high specificity of the phage–host interaction. Finally, since phages are a natural phenomenon, we can affect pathogenic bacteria avoiding harmful chemical agents.
*To obtain ultrathin sections, cells are embedded into a special resin; the resulting hard blocks are cut into sections 60–80 nm thick, using an ultramicrotome equipped with a glass or diamond knife
References
Ackermann H. W., Prangishvili D. Prokaryote viruses studied by electron microscopy. 2012. N. 157. P. 1843—1849.
Ackermann H. W., Tiekotter K. L., Murphy’s law – if anything can go wrong, it will // Bacteriophage. 2012. N. 2:2. P. 122—129.
Bacteriophages methods and protocols / Ed. A. M. Kropinski, R. J. Clokie. Humana Press, 2009. V. 1.
Duckworth D. H. Who discovered bacteriophage? // Bacteriological reviews. 1976. V. 40. N. 4. P. 793—802.
Introduction: a short history of virology // Viruses and man: a history of interactions / Ed. M. W. Taylor. Springer, 2014. P. 1—21.
Krylov V. N. Phage therapy in therms of Bacteriophage genetics: hopes, prospects, safety, limitation // Rus. J. of genetics. 2001. V. 37. N. 7. P. 869—887.
Matsuzaki S., Rashel M., Uchiyama J., et al. Bacteriophage therapy: a revitalized therapy against bacterial infectious deseases // J. Infect. Chemother. 2005. N. 11. P. 211—219.
The photos are by the courtesy of the authors, and drawings, by Zhenya Vlassov













