Mini Nano-Engineers
Remaining together through forming colonies (lower organisms) or tissues (higher organisms) or becoming dispersed as the need arises is one of the fundamental principles of biological existence.
Having taken the evolutionary step of developing a rigid cell wall of two parts, an epitheca and a hypotheca surrounding a polarised cell, the diatom cells were, with very few exceptions, denied any choice in the manner in which they could remain together.
The only method available to them was to form chains if the cells remained attached to each other following cell division...
R. CRAWFORD: Certainly silica is a brilliant “choice” from the point of view of its availability, strength and transparency (remember these are photosynthetic organisms), but it also has to allow the passage in and out of solutes and dissolved gasses. This is a subject that has hardly been touched on yet it is an area of study that is desperately in need of a team of physiologists and physicists. For almost the whole of the history of diatom study we have been aware that the wall is full of holes. In point of fact, what appear in the light microscope as holes have been established from electron microscopy as thin areas (areolae) of the valve in which there are pores of a very much finer dimension and, if anything, even more precisely arranged. It is through these that dissolved nutrients and gasses must pass.
This also brings us to another aspect of the diatoms that has intrigued and fascinated diatomists over the years — their precise organisation. How do diatoms create and reproduce such precision when they form new wall components? As a seminal review of the subject of diatom morphogenesis has pointed out, several processes are expressed at different scales and extend from the gross form which may be brought about by the parent frustule outside the new cell, by larger or smaller vesicles or even mitochondria within the cells or even within the deposition vesicle by the chemically arranged building blocks of the template and for an up-to-date picture of events at this scale. Macro- and micro-processes of diatom morphogenesis are receiving special attention today because of their potential in nanotechnology.
Harmony of beauty and expediency
Remaining together through forming colonies (lower organisms) or tissues (higher organisms) or becoming dispersed as the need arises is one of the fundamental principles of biological existence.
Having taken the evolutionary step of developing a rigid cell wall of two parts, an epitheca and a hypotheca surrounding a polarised cell, the diatom cells were, with very few exceptions, denied any choice in the manner in which they could remain together.
The only method available to them was to form chains if the cells remained attached to each other following cell division...
Everyone is familiar with the common, though misleading, perception that animals move and plants are rooted to the same place. The idea that, for those same plants dispersal is important in the form of pollen, seeds or in some cases vegetative parts of the parent, is also well known. So much for terrestrial systems, but such considerations are also relevant to aquatic environments. Only the vegetative (non-sexual) stages concern us here but, put simply, if a cell divides it can remain attached or it can become separated from its sibling and the two will be carried in different directions. If the cell has successfully grown and divided it usually means that conditions in the immediate environment are good and that it would thus be beneficial for both cells produced at the division not to be separated and to remain where they are. Obviously this is more the case when the parent cell is actually attached to a surface than if it is freely floating. On the other hand, separation of the cells would optimize dispersal, especially in a turbulent habitat, and this would be of clear benefit, for example in conditions of nutrient depletion. The opposite situation where dispersal is avoided is of further benefit to the population because it maximises the chance of encounter of gametes if a population enters a sexual phase.
TRIBOLOGY (from Greek tribos — friction) is the branch of engineering that deals with the interaction of surfaces in relative motion (as in bearings or gears): their design, friction, adhesion, lubrication and wear. Tribology is the science and technology and practice. Recently, micro- and nanotribology have been gaining increasing interest. These fields deal with tribology at size scales of functional elements from 100 micrometers to some tens of nanometers. Frictional interactions in microscopically small components are becoming increasingly important for the development of new products in electronics, life sciences, chemistry, sensors and, by extension, for all modern technology.
The aim of biotribology is to gather information about friction, adhesion, lubrication and wear of biological systems and to apply this knowledge to technological innovation and to the development of environmentally sound products. This new interdisciplinary field of research combines methods and knowledge of physics, chemistry, mechanics and biology. There are many examples of tribology in biology. Surfaces in relative motion occur for example in joints, in the blinking of the eye and in a foetus moving in the mothers womb. Examples for systems with increased friction are bird feather interlocking devices and friction in fish spines
The field of biomicro- and -nanotribology was founded in response to strong needs of technology. Continuous miniaturization of technological devices such as hard disk drives and biosensors has increased the need for the fundamental understanding of tribological phenomena at the micro- and nanoscale. Biological systems excel at this scale and so their strategies may serve as templates for new engineering devices and the rigid parts of diatoms in relative motion and subject to various forces as we have seen above are therefore of particular interest for biomimetic microsystems engineering
It should be noted that among diatoms one can see that chain-forming species exist alongside unicellular ones. Chains are found in several kinds of habitat but chiefly epiphytic (attached to plants), epipsammic (to sand grains), epilithic (to stones or rock) and planktonic (free-floating in the water). In all situations the diatoms may be subject to considerable agitation due to the turbulence of the water and here we shall consider the forces that may be brought to bear on the diatom cell and particularly those cells that are bound together in a chain by means of linking spines.
Like a key to a lock
Three kinds of attachment may be found in diatoms.
The cells may be attached by adhesive material that is passed to the outside through tubes in the cell wall. In a very small number of genera the siliceous structures of sibling valves may be fused. In many other genera the sibling valves are linked during formation by interlocking spines of varying complexity and size, fitting to each other like the key to the lock. Those structures are generated while sibling valves are forming.
“Producing each of its creations... nature intermingled the harmony of beauty and the harmony of expediency and shaped it into the unique form which is perfect from the point of view of an engineer” (M. Tupolev)
The design of these spines is often more than a mechanism constructed to keep cells from drifting apart and this prompted us to examine the forces acting on the cells in a chain. The forces are as follows. 1. the tension pulling cells apart; 2. the force pushing them together and 3, the rotational force or torque tending to twist the cells in the chain. When considering the cells of a chain in the varyingly turbulent environment, for example subjected to inshore waves on an exposed shore, it is clear that a cocktail of forces 1, 2 and 3 will be felt. Forces 1 and 2 will have been taken for granted by anyone giving superficial thought to life in a chain but perhaps not force 3.
We examined a number of chain-forming diatoms with this force in mind and have reached the conclusion that many diatoms have had to deal with it from very early times in diatom evolution.
It was found out that many fossil species over 30 million years old keep the two valves together and prevent it twisting. Ellerbeckia is an extreme example.
The engineering solutions of Ellerbeckia
The genus Ellerbeckia is remarkable for having a very long fossil record, a very early separation in diatom evolution from other genera and an extremely close linking of the sibling valves.
The cell wall is very robust and the valve face and mantle (the side of the cell) are at right angles. If we look at the valve face of a fragment of the interlocked sibling valves we can see how intimately the features of one interlock with those of the other. In some species linking structures are found towards the middle of the valve face but visible here is a feature common throughout the genus ridges on one valve fitting into depressions on the other. If we examine linked cells from the side — the girdle view — we see that the spines are so closely interlocked that the two cells will not pull apart unless the spines are broken. Clearly with such linking structures the connection between two cells will neither be twisted nor moved together or apart very much but this leads us to another problem.
If the cell to cell connection is now safe from destruction the junction between valve and girdle bands will now be the weakest part of the chain and the most vulnerable to the various forces. Here Ellerbeckia shows us a unique character among diatoms. In all other genera yet recorded the interface of valve and first girdle band (valvocopula) is smooth. In most genera, especially in the “modern” motile unicellular diatoms, the valvocopula does not have a very close or precise relationship with the younger of the two valves, the hypovalve, but in Ellerbeckia all three components, hypovalve, valvocopula and epivalve, are locked very tightly and relate to each other as do two fists of a hand placed knuckle to knuckle but with the girdle band between them.
The result of such a close connection at both of the potentially vulnerable sites might have one of two consequences. Either the chain is as solid a rod as if it were a string of fused cells (although this would increase the possibility of breaking the colony at high impact loads) or a certain amount of the energy transferred into the colony by the action of the forces (i.e. the work done on the chain) is dissipated through all of the small, but close, links between the components. It would seem that the whole system functions as one because we do not find chains with the occasional linking structure broken.
Why is it that in this genus we find a character that is found nowhere else among the multitude of diverse forms among the diatoms? Why do we not find a more secure valve/girdle/valve junction in ANY other diatoms? How is it that other diatoms, particularly chain forming species exposed to similar forces manage to avoid destruction? The answer is probably in the evolution history of this old genus formed in shallow turbulent seas of the past. The fact that other, quite different genera thrive just as well alongside Paralia and Ellerbeckia adds to the mystery but is at least testament to the great diversity of the diatoms and to their thrival strategies. This diversity may prove to be a rich vein of discovery for the biotechnology of the future.
Building of silica
To make the walls of this house, diatoms take up a dissolved form of silicon (silicic acid) from the environment and during the period of cell wall formation control its polymerization to form structured silica with features on the nanometer to micrometer scale. An analogy can be made between formation of the diatom frustule and construction of a building. Not only does one need to have the appropriate equipment to make various structures in a building, but also a coordinated supply of raw materials to meet the needs at different stages of building construction. This not simple process in diatoms is provided by special cellular transport proteins.
Silicic acid and silicate refer to the soluble forms of silicon, Si(OH)4 and SiO(OH)3- respectively.
Polymerized silicon is called silica, which itself can exist in different structural forms
At 97%, silicic acid is the dominant form of silicon in the ocean. The remaining 3% is comprised of silicates. Because diatoms require silicon for their cell walls, and because they are extremely abundant, they have greatly reduced the amount of silicon in the world’s oceans. Prior to the evolution of diatoms and other siliceous organisms, the world’s oceans were nearly saturated with silicon (1.7-2.5 mM). Today, that number is estimated to be <70 µM globally, but can be <10 µM and sometimes <1 µM in surface waters where silicifying organisms dominate.
Inside the diatom cell, concentrations of silicic acid have been measured between 19 and 350 mM, depending on the species. This poses two main problems. First, because extracellular concentrations of silicic acid can be over one thousand times lower than intracellular levels, diatoms must take up silicic acid against a very steep concentration gradient. In our building analogy, it would be like a heavily loaded concrete truck having to climb a very steep hill to get through the gate of the building site. Second, the chemical nature of silicic acid is such that it autopolymerizes into silica at concentrations above 2 mM. This would be highly detrimental if it happened in the wrong location in a diatom cell; in our analogy, it would be like a concrete truck entering the construction site but pouring the concrete in the yard instead of taking it to the appropriate location in the building. Because we do not observe silica polymerization throughout the diatom cell, diatoms must have a mechanism of maintaining supersaturated levels of silicic acid in an unpolymerized, soluble form, analogous to how concrete is maintained in its liquid form by continuous mixing. While this mechanism has not yet been identified, the limited data available suggest it may occur through the association of silicic acid with organic silicon binding components or proteins.
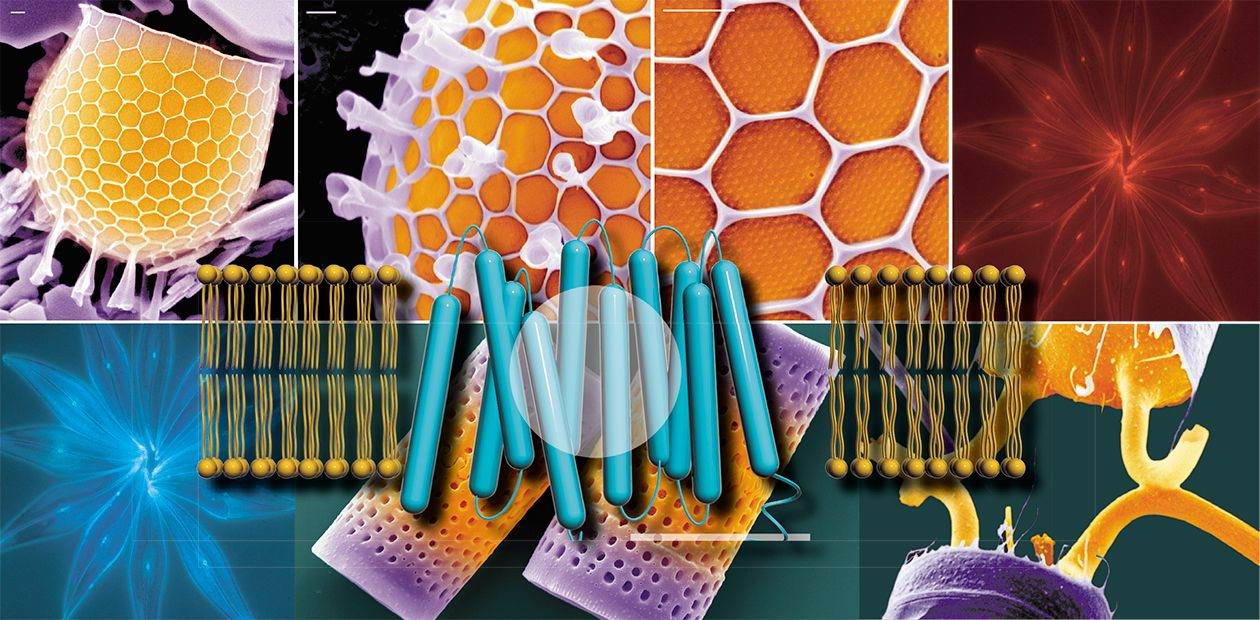
In order to support their requirement for silicon, diatoms possess proteins designed to specifically recognize, bind, and transport silicon from the environment. These proteins were originally identified indirectly by measuring silicon uptake into diatom cells. The proteins are called silicon transporters, or SITs, and are membrane-spanning proteins that form a channel enabling the specific passage of silicic acid across the normally impermeable lipid bilayer membrane. It was shown that silicon uptake was coupled to the presence of extracellular sodium. Because extracellular sodium is high, a favorable concentration gradient exists for its uptake, and SITs can transduce the energy generated by sodium uptake into movements that enable co-transport of silicic acid. Thus SITs are sodium/silicic acid symporters.
Genes encoding SITs were first identified in the marine pennate diatom, Cylindrotheca fusiformis. Five distinct genes were identified, each with a different level or pattern of mRNA expression during cell wall formation. Recently, genome sequencing of the centric diatom, Thalassiosira pseudonana, led to the identification of three SITs. It has been also been suggested that SITs have varying affinities or transport capabilities, meaning some SITs may function primarily under conditions of low or high silicon. Alternatively, SITs may function as silicon sensors to detect the amount of silicon available to the cell, or may function to transport silicic acid into the silica deposition vesicle.
SITs were the first proteins shown to specifically and directly interact with silicic acid, and although other silicon-interacting proteins have since been identified, SITs remain the only proteins known to interact with soluble silicon without catalyzing its polymerization to silica. Sequencing of SITs revealed that they are unique and comprise a completely new family of transporters. While this makes them both intriguing and mysterious, it also makes it difficult to study the details of their function because there are no homologues for modeling them.
Two models have been proposed for the mechanism by which SITs co-ordinate silicic acid. Both of these models are based on sequence comparisons and limited experimental data, and although they propose different mechanisms for SIT function, they provide, for the first time, hypotheses that researchers can test in laboratory experiments.
Two alternative models have been proposed for the mechanism by which SITs coordinate silicic acid.
In the first model a conserved sequence motif (CMLD) serves as the silicic acid binding site of SITs via coordination with zinc. This motif is found not only in a majority of diatoms but in much older Chrysophyte (golden) algae (Likhoshway et al., 2006).
The second model is based on conformational change in the transporter (SITs). Comparison of 29 partial SIT sequences from 12 diatom species led to the identification of a conserved sequence motif (GXQ) identified in four distinct regions of the protein. These motifs are proposed to be responsible for coordinating silicic acid through hydrogen bonding (Thamatrakoln et al. 2006)
The tools of molecular biology can be applied to specifically modify amino acids proposed to be involved in transport, as a means to directly test their involvement. An important tool to be developed in the further characterization of the SITs is to express genes encoding these proteins individually in a host cell other than a diatom. This is because multiple SITs are present in all diatom species examined, therefore one cannot study an individually modified SIT when other unmodified SITs are present.
In addition to their essential roles in diatom silicon metabolism, another reason for studying SITs is that they provide a model system to understand in general how biological components interact with silicon. In addition to silicon’s role in diatoms and sponges, it is a highly beneficial element in plants, where it contributes to rigidity, productivity, and disease resistance, and in vertebrates, where it is an essential element for formation of bone, cartilage, and connective tissue. Understanding the details of how SITs recognize and bind silicic acid, the mechanism of transport across the membrane, and the ways by which SITs are regulated could provide essential insights into silicon’s involvement in other life processes