The Ribosome: A Minifactory for Protein Production
Synthesis of proteins, the most important structural and functional “bricks” of any organism, is perhaps among the most intricate processes occurring in living beings. A genuine understanding of the molecular processes underlying protein synthesis could shed light on incredibly ancient events connected with the mystery of the origin of life itself...
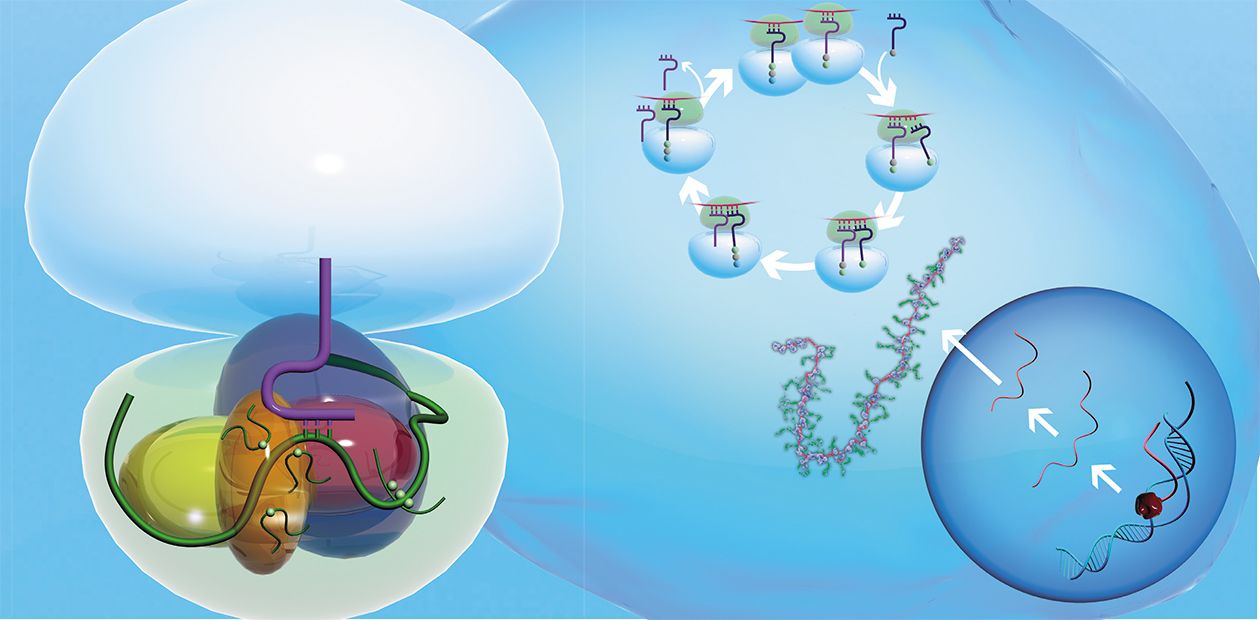
Proteins in all the living organisms from the simplest bacteria to humans are synthesized by the specialized cell devices — ribosomes. In these unique factories, the protein chain is formed of individual amino acids.
Ribosomes are abundant in the cells that are involved in an intensive protein synthesis: for example, one bacterial cell contains about 10,000 minifactories, constituting up to 30 % of the cell dry weight! The cells of higher organisms contain fewer ribosomes, whose number depends on the type of tissue and level of cell metabolism.
The ribosome synthesizes protein at an average rate of 10–20 amino acids per second. The accuracy of translation is exclusively high — an erroneous insertion of an “incorrect” amino acid residue into a protein chain occurs on the average in one amino acid per three thousand residues (at a mean length of a protein chain for humans equaling 500 amino acid residues), i. e., only one error per six proteins.
On the Genetic Code
The program specifying the sequence of amino acid residues in proteins is recorded in the cell genome: it was discovered about half a century ago that the amino acid sequences of all the proteins are directly encoded in DNA with the help of the so-called genetic code. According to this code, which is universal for all the living organisms, a certain codon — triplet of nucleotides, which are elementary units of the DNA chain, corresponds to each of the 20 existing amino acids. Any protein is encoded in DNA by a sequence of codons. This sequence is called a gene.
One cell can contain up to 10,000 ribosomes, protein minifactories, which compose up to 30% of the cell dry weight.Well, how is this genetic information transferred to the ribosome? The chain of another informational molecule, ribonucleic acid (RNA), is synthesized from an individual gene as a template. This process of copying the gene, named transcription, is performed by special enzymes — RNA polymerases.
However, the RNA thus synthesized does not yet represent the template for protein synthesis: certain “noncoding” fragments of nucleotide sequence must be excised from it, especially, in the cells of eukaryotes (organisms with a formed nucleus). This process is named splicing.
The accuracy of protein synthesis by the ribosome is exclusively high — the error rate in humans amounts to one “incorrect” amino acid per three thousand residues (or six proteins)Thus, the messenger RNA (mRNA) is produced; this mRNA is used by ribosomes as a program for synthesizing proteins. The synthesis itself, i. e., the rendering of genetic information from the language of mRNA nucleotide sequence into the language of amino acid sequence of protein is called translation.
Decoding and Synthesis
In eukaryotic cells, one mRNA is usually translated by numerous ribosomes, which form the so-called polysomes, distinctly visible with the help of electron microscopy, allowing tens of thousands — fold magnification.
How are amino acids, the structural bricks for protein synthesis, conveyed to the ribosome? As long ago as in the 1950s, the specialized “carriers” transporting amino acids to the ribosome were discovered; they are short (with a length not exceeding 80 nucleotides) transfer RNAs (tRNAs). A special enzyme attaches an amino acid to one of the tRNA ends; note that a strictly determined tRNA corresponds to each amino acid. The protein synthesis on ribosome comprises three main stages: initiation, elongation of the polypeptide chain, and termination.
The ribosome itself is one of the most intricately organized molecular machines of the cell; it consists of two unequal parts, the so-called subunits (small and large). It is readily dividable by centrifugation, at superhigh speeds, in special tubes filled with the solution of sucrose at concentrations increasing from top to bottom. As the small subunit is half the weight of the large subunit, they move from top to bottom of the tube at different speeds.
The small subunit is responsible for the decoding of genetic information. It consists of a high-molecular-weight ribosomal RNA (rRNA) and several dozens of proteins (about 20 protein species in prokaryotes and over 30 in eukaryotes).
In tumor cells, the level of certain ribosomal proteins increases drastically. A possible reason is a failure in the mechanisms underlying the self-regulation of their productionThe large subunit, responsible for the formation of peptide bonds between amino acid residues, comprises several rRNAs: one high-molecular-weight and one (or two in the case of eukaryotes) low-molecular-weight rRNAs and, similarly, several dozens of different proteins (over 30 in prokaryotes and up to 50 in eukaryotes). The following figures allow us to assess the scale of the ribosome activity: the ribosome RNA constitutes about 80 % of the total cell RNA; the tRNA, which conveys amino acids, makes up about 15 %; whereas the messenger RNA, carrying information about the protein sequence, constitutes only 5 %.
Note that the ribosomal proteins have manifold additional functions, which they fulfill at various stages of the cell life activity. For example, the human ribosomal protein S3, one of the key proteins forming the ribosomal mRNA-binding center, is also involved in the “repair” of DNA damages (Kim et al., 1995), apoptosis (programmed cell death; Jung et al., 2004), and the protection of a heat shock protein from destruction (Kim et al., 2006).
In addition, an excessively intensive synthesis of some ribosomal proteins may indicate a development of cell malignant transformation. For example, a considerable increase in the levels of five ribosomal proteins was detected in tumor cells of large intestines (Zhang et al., 1999). Recently, researchers from the Laboratory of Structure and Function of Ribosomes, Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, discovered a new self-regulation mechanism involved in the biosynthesis of human ribosomal proteins which is based on the principle of feedback. An uncontrolled synthesis of ribosomal proteins, characteristic of tumor cells, most likely results from a failure of this particular mechanism. Further research in this field is of special interest not only for scientists, but also for physicians.
Works as a “ribozyme”
Amazingly enough, the secondary structures of prokaryotic and eukaryotic ribosomal RNA of small and large subunits are very similar despite billions of years of evolution that separate bacteria and humans.
Until recently, information about the packaging of rRNA in subunits and its interaction with ribosomal proteins was scarce. A revolutionary advance in our understanding of the ribosome structure on molecular level occurred on the eve of the new millennium, when x-ray structure analysis allowed the structures of protozoan ribosomes and their model complexes with mRNA and tRNA to be determined at the level of individual atoms. This formed the basis for understanding the molecular mechanisms involved in the decoding of genetic information and in formation of bonds in the protein molecule.
It appeared that both most important functional centers of a ribosome — the decoding center in the small subunit and the center responsible for synthesis of the protein chain in the large subunit — are formed not of proteins but of ribosomal RNA. This means that the ribosome operates similarly to ribozymes, unusual enzymes composed of RNA instead of proteins.
Nonetheless, ribosomal proteins also play an important role in the work of ribosomes. In the absence of these proteins, ribosomal RNAs are completely incapable of either decoding genetic information or catalyzing the formation of peptide bonds. The proteins provide a complex “packaging” of rRNA in the functional centers, which is necessary for the work of ribosomes; serve as “transmitters” of the changes in ribosome spatial structure, necessary during its operation; and bind various molecules influencing the rate and accuracy of protein synthesis.
The working scheme of the protein cycle is principally the same for the ribosomes of all living beings. However, it is still unknown to what degree the molecular mechanisms involved in the operation of ribosomes are similar. The most important insufficient information is related to the organization of ribosomal functional centers in higher organisms, which are essentially worse studied as compared with protozoan ribosomes.
This is because the numerous methods that have been successfully applied to the research into prokaryotic ribosomes have appeared inappropriate for eukaryotes. For example, it is impossible to obtain eukaryotic ribosomes in a crystalline form suitable for x-ray structure analysis, and to “assemble” their subunits in tubes from a mixture of ribosomal proteins and rRNAs, as is easily performed with protozoan ribosomes.
From Lower to Higher
Nonetheless, there are methods to obtain data on the structure of ribosomal functional centers in higher organisms. One of such methods is chemical affinity cross-linking, developed 35 years ago at the Department of Biochemistry with the Novosibirsk Institute of Bioorganic Chemistry, USSR Academy of Sciences (now, Institute of Chemical Biology and Fundamental Medicine) under the guidance of Academician D. G. Knorre.
This method utilizes the short synthetic mRNAs carrying at a specified position the chemically active (“cross-linking”) groups that can be activated at a particular moment (for example, by irradiating with a soft ultraviolet light).
The method of chemical affinity cross-linking was designed 35 years ago at the Department of Biochemistry (Novosibirsk Institute of Organic Chemistry, Siberian Branch of the USSR Academy of Sciences, (now, Institute of Chemical Biology and Fundamental Medicine SB RAS) under the guidance of Academician D. G. Knorre. Before x-ray structure analysis was applied to ribosomes, this method was used worldwide for studying prokaryotic ribosomes.Today, this method remains the main approach to the research into the structure — function organization of ribosomes of higher organisms
The advantage of this method is that the cross-linking group can be attached to virtually any nucleotide residue in mRNA to obtain detailed information about its neighborhood in the ribosome. Using a set of short mRNAs with different locations of the cross-linking group, we succeeded in determining the ribosomal proteins and the rRNA nucleotides in the human ribosome that formed the channel for reading genetic information during the translation process.
It was demonstrated for the first time that all the nucleotides of the human small subunit rRNA adjacent to the codons of mRNA were localized to relatively conservative regions in the secondary structure of rRNA molecule. Moreover, their location coincided with the positions of the corresponding nucleotides in the secondary structure of the rRNAs from lower organisms. This suggested that this part of the small subunit ribosomal RNA formed an evolutionarily conservative “core” of the ribosome, structurally identical in all the organisms.
On the other hand, a number of principal distinctions have been discovered between the structures of mRNA-binding channel in the ribosomes of humans and lower organisms. It appeared that the ribosomal proteins of higher organisms played a considerably more important role in formation of this channel as compared with prokaryotes; moreover, certain proteins that are involved in this process lack any “twins” (homologues) in the lower organisms.
Thus, why have specific features in the organization of the ribosomal decoding center of higher organisms appeared during the evolution, despite the fact that the function of ribosomes have not virtually changed? Presumably, this is connected with a more complex and multistage regulation of protein synthesis in eukaryotes compared with prokaryotes, when the ribosomal proteins of mRNA-binding channel may interact not only with mRNA, but also with other factors influencing the efficiency and accuracy of translation. Further research will demonstrate whether this hypothesis is true.