Biochips: Diagnosis Is a Matter of Technique!
What are biochips? Today, this buzzword means the miniature devices for determining specific interactions between biological macromolecules. Nucleic or ribonucleic acids (including genomic DNA fragments), proteins, and other molecules can serve as probes in such chips.
Over a short time period, biological microchips have formed a separate field of analysis with manifold applications, from merely practical uses in medicine, pharmacology, ecology, and forensic examination to basic research, including molecular biology and molecular evolution...
The other name for biochips, microarrays, most precisely defines their essence, as these devices are actually molecules orderly arranged on a solid carrier surface. As a rule, a plate of glass or plastic (or, sometimes, of other materials, for example, silicon) is used as a carrier. In this respect, biological chips are similar to electronic chips, which are manufactured on silicon plates.
As biological chips are usually small, they are most frequently called microchips. Nonetheless, a tremendous number of various probe molecules can be placed on the small area of a biochip. After the molecules tested interact with the chip, the information contained on this area is read with a special device.
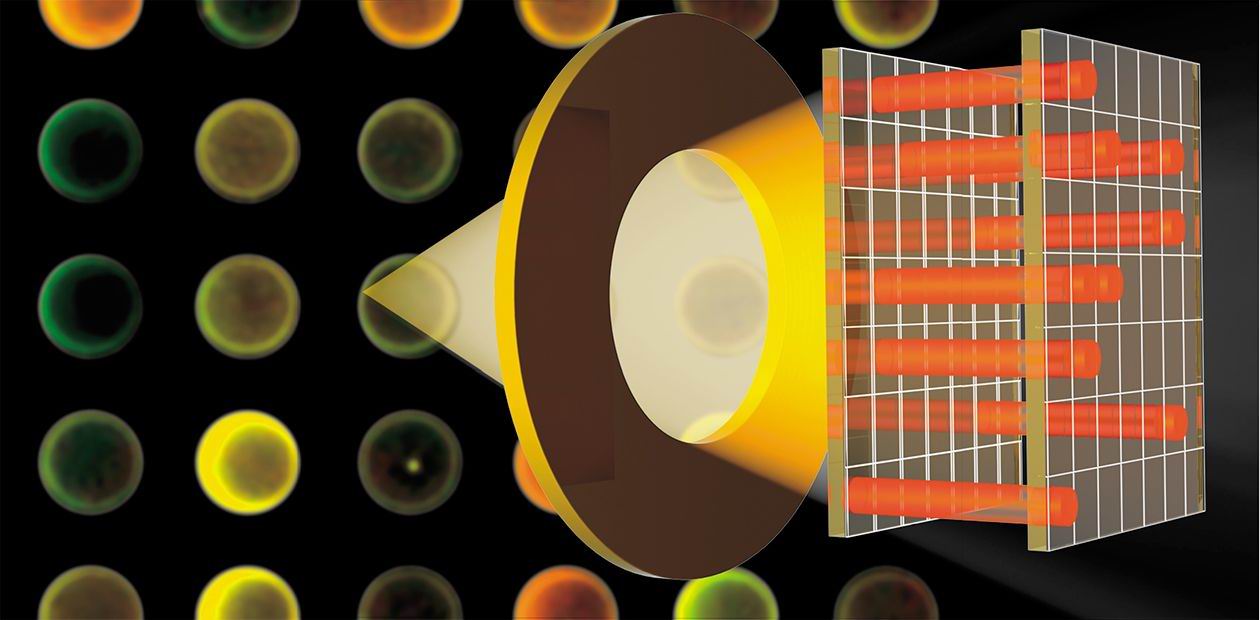
The essence of microchip diagnostics is recording the event of the so-called ligand – receptor interaction. One of the interacting partners — ligand, or receptor — is immobilized on a tiny, yet stringently specified, surface of the microchip called spot. (In some cases, the surface of microchips is structured as microcells (nanoplates) or microelectrodes.)
Each spot is composed of identical molecules. When analyzed, the tested sample is placed onto the microchip to record the event of its interaction with the molecules contained in its certain spots. The result of such interaction is a characteristic pattern of the interaction between the sample and the microchip.
Fluorescence is frequently used for recording the interaction between a receptor and ligand. For this purpose, the analyzed sample is labeled with a fluorescent compound before the incubation with a microchip. After the incubation, the microchip is scanned with a laser scanner to read the fluorescence intensity distribution on its surface. Thus, the spots with the molecules that have reacted with the sample studied are detected.
If the molecular complementarity conditions, which determine the correspondence between immobilized molecular probes and dissolved labeled molecules analyzed, are met, the complexes formed will be most stable thermodynamically. Consequently, such complexes will be more abundant at certain temperatures than the complexes formed due to incomplete complementarity, and the fluorescence signal of the former will be more intensive.
Studying the profiles of… expression
Today, the technology of oligonucleotide* biochips is the best developed, and the most widely applied among them are the so-called expression chips.
To understand how they work, let us recollect how the vital activities of our cells are regulated. An intricate network of molecular interactions controls the processes of intracellular metabolism, cell division, and differentiation; this network determines the level of expression (functioning) of various genes. The concentration of the corresponding messenger RNA (mRNA), which is synthesized on the template of DNA, the carrier of hereditary information, reflects the expression level. The dynamics of changes in the concentration of a certain mRNA indicate that the corresponding gene is switched on or off, while considerable deviations of this characteristic from the norm may suggest the occurrence of certain diseases.
It is possible to manufacture biological chips by various methods. Molecular probes attached to the carrier surface of a microchip can be either synthesized directly on the microchip starting from the first link covalently bound to the surface or beyond the microchip to be immobilized on the surface later. The former method is used when producing the chemical “libraries”, where short oligonucleotides or peptides are used as probes; whereas the latter is applied for immobilizing extended polynucleotides, proteins, or other large receptor molecules.
Affymetrix (USA), one of the largest companies involved in manufacturing microarrays, uses the production design created for electronic chips. The Affymetrix microarrays are produced on glass plates by photolithography using specialized micromasks. Application of the well-optimized methods of electronic industry have led to impressive results: one chip of the size of only several microns can contain millions of various spots (the regions containing the same molecular probes)
The diversity observable in gene expression profiles is in the focus of researchers’ attention due to their biological and clinical importance. From this standpoint, microchip technologies have appeared very useful in studying such an intricate disease as cancer. This technology makes it possible to monitor the expressions of tens of thousands of genes simultaneously, thereby obtaining a kind of molecular “portrait” of a cell. Today, it is already possible to diagnose many kinds of cancer by examining gene expression profiles in the cell, as well as to determine the risk groups and forecast the disease development.
At present, diagnosing cancer requires a joint examination of a patient by expert physicians — oncologists, pathologists, and cytogeneticists. However, the final conclusion can vary considerably depending on various subjective factors, such as the level of their expertise and the particular methodological approaches used. Application of corresponding microchips makes it possible to do without many experts and to elevate considerably the accuracy of disease diagnosis and prognosis.
The expression biochips available utilize the immobilized DNA of two types: first, relatively short synthetic DNA fragments with a length of 20–60 nucleotides, which correspond to particular regions of a gene (in this case, each gene is characterized by 20–40 spots containing distinct probes); and, second, large fragments of coding DNA with a length of 100–600 nucleotides obtained by using the corresponding enzymes (the so-called DNA microchips).
The procedure of preparing a sample for microchip-based assay includes isolation of total RNA, which is used as a template for synthesizing the strands of coding DNA with the help of reverse transcription reaction. The enzyme T7 RNA polymerase makes it possible to produce many copies of RNA from each molecule of such cDNA. As a rule, the synthesized RNA molecules are concurrently labeled with a fluorescent dye (using labeled nucleotides in the synthesis).
When working with oligonucleotide microchips, researchers routinely use fluorescent labels of only one type for labeling the sample of synthesized RNA; they determine the levels of gene expression by comparing the obtained fluorescence signals with the signals from internal control points of the microchip.
However, when working with the microchips involving cDNA, one usually needs two labels, namely, the control sample (taken from the normal cell) is labeled with one fluorescent dye and the analyzed sample (for example, of a cancer tumor), with another; then they are mixed and hybridized to the same microchip. The ratio of the signals from two different labels at each spot of the microchip demonstrates an increase or decrease in the expression of a particular gene during the corresponding disease.
Disease, evolution, crime…
A single experiment with biochips gives data on expression levels of tens and hundreds of thousands of genes. Rather complex mathematical tools, first and foremost, cluster analysis, are used to process this large volume of information.
Such analysis detects groups of genes where changes in the expression level indicate the presence of a particular disease. The further diagnosing of patients is based on the analysis of these genes. In some cases, it is possible that the disease previously diagnosed as a certain cancer type actually is a combination of several concurrent diseases.
Expression microchips make it possible not only to study the molecular mechanisms underlying various diseases, but also to test the effects of various drugs on patients (determining the indications and contraindications of the drugs individually) and evaluate the organism’s response to the action of manifold external factors.
In addition, oligonucleotide microchips find a wide application for analyzing genomic polymorphism (diversity). It is known that the genomic DNA sequences of individual living organisms are, as a rule, somewhat different from the sequences compiled in databases. These differences are mainly due to point mutations, insertions, and deletions (loss) of nucleotides. In their turn, such differences in the carrier of hereditary information lead to the intraspecies diversity of organisms, and the accumulation of these differences, to divergence between species. Thus, the study into genomic polymorphism with the help of microchips can enhance the clarification of a most important mechanism involved in molecular evolution.
Another field of application for the biochip technology is the study of mutations in genomes of various pathogens, which allows to determine precisely the species of bacteria or viruses. For this purpose, extended fragments of genomic DNA or RNA of a pathogen are hybridized to the microchip containing special oligonucleotide probes.
In particular, our Lab in collaboration with the Laboratory of Method Development, Food and Drug Administration (Washington DC, USA) proposed a microchip-based method for detection and classification of the orthopoxviruses pathogenic for humans. Variola and monkeypox viruses are the orthopoxviruses most dangerous for humans. Cowpox, camelpox, and vaccinia viruses are less hazardous yet capable of causing human diseases. Although variola virus was eradicated in the 1970s, the probability of its reemerging in the human population cannot be excluded. The potential source for reemergence of this infectious agent can be either opened graves of smallpox cases or deliberate bioterrorism attacks. As the main part of the world population lacks the immunity to this virus, infection with smallpox could have catastrophic consequences.
The microchip that we have designed is able to diagnose the orthopoxvirus species pathogenic for humans. Moreover, the clinical presentation of orthopoxvirus-infected cases is very similar to that caused by herpesviruses types 1, 2, and 3. Our microchip can distinguish between the patients infected with orthopoxviruses and those infected with herpesviruses. This is of utmost importance, as the diseases in question require different therapies despite the similarity in clinical manifestations.
A microchip for the diagnosing of retinitis pigmentosa, a complex eye disease causing blindness, was designed at the Kellogg Eye Center with the University of Michigan (USA). The arRP-1 microchip, developed at the same Center, makes it possible to detect the 180 mutations in nucleotide sequences of 11 genes that cause degeneration of retina during this disease.
Of special interest are the studies of the mutations in the genomes of pathogenic bacteria that cause antibiotic resistance. For example, approximately 100,000 people annually get tuberculosis in Russia; all over the world, this disease takes away about three million lives a year. The most dangerous aspect of this situation is quick spreading of the tuberculosis forms resistant to standard antibiotics of rifampin type.
That is why an early diagnostics of tuberculosis and detection of antibiotic-resistant tuberculosis agents are so important. For this purpose, the Engelhardt Institute of Molecular Biology created the diagnostic variants of biochips for express diagnostics of tuberculosis combined with the assay for drug resistance. The assay time is essentially decreased, from 60 days (standard assay) to 1 day. This allows the expedient indication of reserved therapeutics to the cases with drug-resistant tuberculosis variants their number exceeds 10 %.
Biochips can find practical application in the field rather distant from medicine, namely, in forensic practice for personal identification. In this case, the nucleotide sequences of the most variable regions of human mitochondrial DNA are used for analysis.
Along with DNA microchips, protein microchips have become widely known; in this variant of biochips, peptides or whole proteins play the role of molecular probes. These microchips make it possible to study protein–protein, protein–DNA, and other interactions. The protein microchips containing the antibodies for specific diagnostics of pathogens are designed most frequently.
There are also other types of microchips, for example, chemical microchips, which contain real combinatorial “libraries” of small chemical compounds. Such microchips provide a simultaneous screening of thousands of potential drugs. Another type, the microchips containing specimens of biological tissues, is used for concurrent analysis of thousands of tissues for determining the protein contents in healthy and pathologically altered tissues and assessing the potential targets for drugs.
Thus, what is the overall significance of introducing biochips into scientific and medical practice? Undoubtedly, the development of new technologies will lead and has already led to unprecedented qualitative advance in these fields.
This is no wonder, as these miniature devices containing millions of spots with most diverse molecular analyzers are able in principle to monitor simultaneously the work of all human genes. These devices open up wide horizons for the research directed to a deeper understanding of the basic functions of organisms in healthy and pathological states and to design fundamentally new drugs. Our health and the health of future generations are in many respects “just a matter of technique”, and the fact that new technologies are available already now causes optimism.
*Oligonucleotides are relatively small DNA fragments with a length of several dozennucleotides.