Nucleic Do-It-Yourself
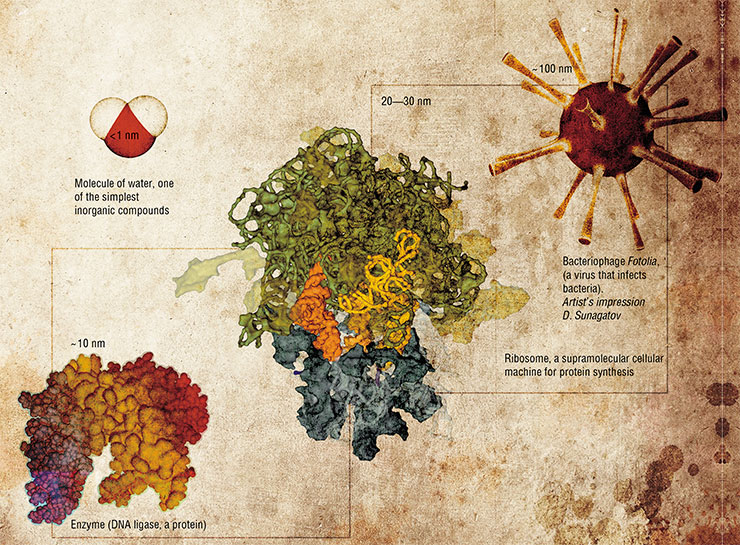
Molecules of relatively simple chemical compounds have the characteristic sizes below one nanometer. However, nanoobjects also include biomacromolecules, such as proteins and nucleic acids, as well as larger molecular “cellular machines” and even viruses, the deceptively simplest living organisms. Basic studies of these objects have always been the main scope of molecular biology. At the same time, scientists worked on the problems of synthesis and applications of natural macromolecules and their artificial analogs, long before nanotechnologies were announced as a state priority direction of research
Once upon a nanotime there were nanopeople
in a nanoland who built nanohouses out
of nanobricks scattered all over
the information field.
Nanofolk nanotale
Nanotechnology is a modern approach to using the properties of materials determined by their nanometer-scale structural elements. Plainly speaking, on transition from macro- or micro- to the nanoscale, the observed physical, chemical, and biological properties of the material may change suddenly. Nanotechnology, or, more strictly, its theoretical branch called nanoscience is concerned with the underlying causes of such quantum leaps in the properties of materials.
From the practical point of view, the existence of “nanomaterial” phenomena allows developing technologies for rational changes of the properties of materials through their specific structuring at the nanoscale. Nanotechnology is capable of producing materials with unusual properties, intricate ultrasmall constructions, and complex mechanisms invisible to the bare eye but uniquely capable of performing operations at the microscopic scale.
Natural complex organic molecules and supramolecular complexes are formed by self-assembly from simpler molecules.Bionanotechnology aims at uncovering the principles that govern the interactions of such structures,with the ultimate goal of reproducing the process of self-assembly in an artificially created system.
This would open the way to the in vitro synthesis of various biological structures, from single enzymes such as DNA ligase to ribosomes and virus-like particles, and even to the design of microscopic biological objects with the desired properties that do not exist in nature
One nanometer (i.e., one millionth of a millimeter) is the size of a simple molecule, while the simplest molecules are even smaller. The main pieces of the bionanotechnological construction kit are much larger organic molecules and their supramolecular complexes, the multipart assemblies of individual molecules. Their size varies from several nanometers to tens or hundreds of nanometers.
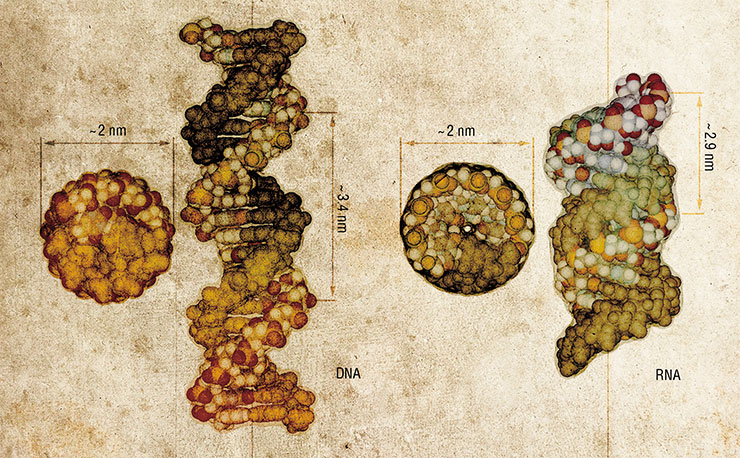
The role of strips, plates and girders in a bionanoconstruction kit is played by molecules of nucleic acids, DNA and RNA. These biopolymers have an amazing ability to self-assemble into distinctive three-dimensional shapes–double-stranded structures held together by complementary interactions. As a result, nucleic acids can be used not only as storage of genetic information (encoded in the sequence of the biopolymer by four “letters” called nucleotides), but also as convenient nanoscale construction blocks.
Nowadays, nucleic acids are almost dirt cheap because efficient methods have been developed for their synthesis. Special automatic synthesizers can churn out nucleotide strands up to a hundred nucleotides long. As the assortment of such molecules is limited only by the ingenuity of the nanodesigner, the number of applications of nucleic acids, an accessible and versatile instrument, is rapidly expanding.
Masks to motors
The simplest example of self-assembly of nucleic acids into nanostructures is formation of DNA-concatemers, polymeric structures formed from blocks consisting of only a pair of oligonucleotides (DNA fragments). This approach can also be used to create two- and three-dimensional structures. In addition to the simple unmodified oligonucleotides, the flat and three-dimensional constructions also employ their conjugates with other inorganic or organic nanoobjects, such as protein molecules.
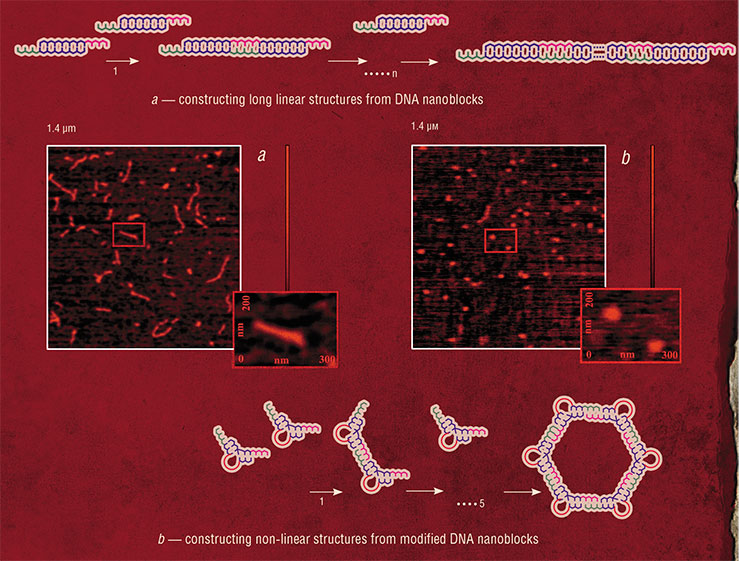
One possible application of this approach is in the production of ordered films, in which inorganic nanoparticles act as branching nodes. These materials would be invaluable for creating self-assembling masks such as those used in modern semiconductor industry to manufacture integrated circuits. In turn, three-dimensional nanoconstructs may serve as unique biocompatible containers for drug storage and addressed delivery to the target organs.

Yet another example of new materials is oligonucleotide conjugates with quantum dots, semiconductor nanoparticles that can fluoresce in the visible spectrum. The sets of oligonucleotide probes labeled in this way simplify a number of problems in biological research (for example, they can be employed to detect several processes in a living cell in parallel), and have also found their use in medical DNA diagnostics and computer tomography.
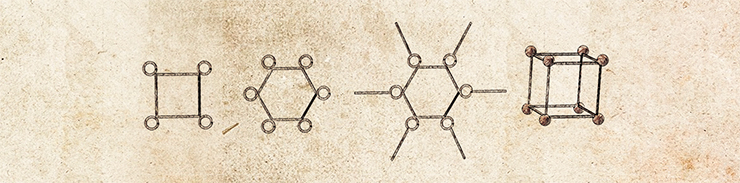
Nucleic aids can also offer a way to construct the so-called cellular molecular machines, or bionanomotors, the devices capable of autonomous movement while transforming the energy of chemical reactions into mechanical work. Autonomy is a very important feature of molecular nanomachines. The host of possible applications for nanomotors includes information processing, controlling chemical reactions, and molecular assembly in various nanoelectronic devices and biosensors.
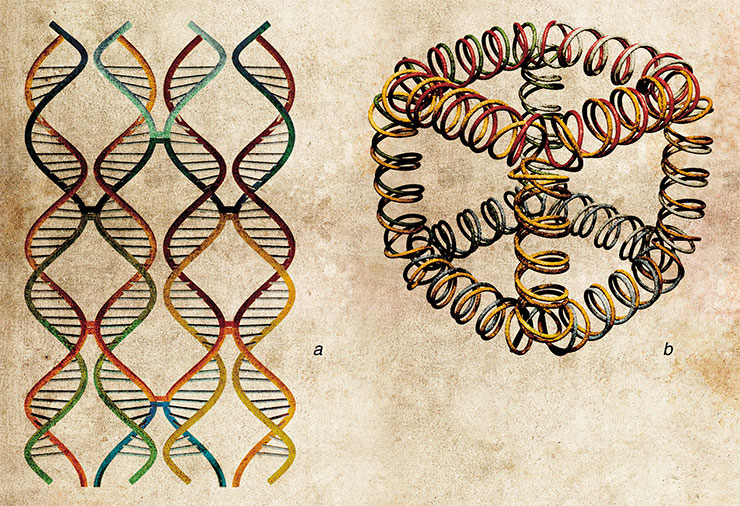
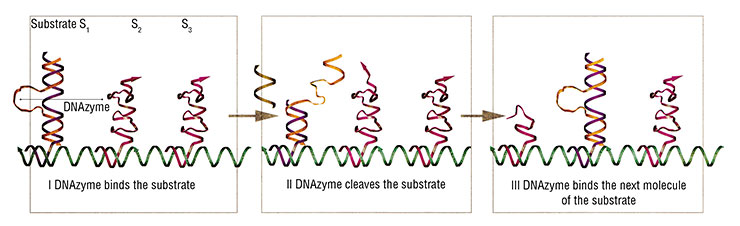
In 2004, a DNA nanomotor was designed based on the 10–23 DNAzyme, one peculiar enzyme made of DNA. These “motors” can perform mechanical movements until they exhaust the “fuel”, their RNA substrate. Although these nanodevices are fully autonomous, their work can be manually controlled, for instance, by adding or depleting the substrate fuel, just as one would step or ease on the gas in the car. Moreover, the nanomotor can be stopped and re-started again using specially designed brake and removal strands.
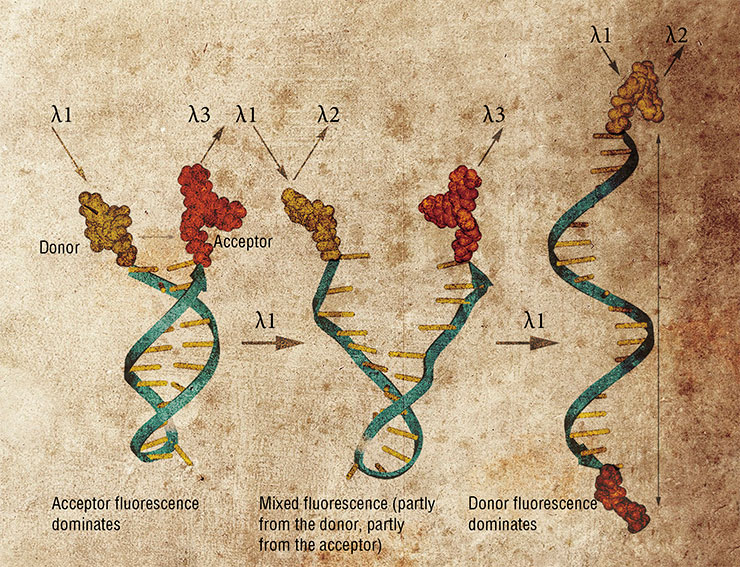
A year later, an even more complex nanomotor powered by 10–23 DNAzyme was made. It is capable of autonomous movement in a direction determined by an oligonucleotide strand track. Such systems in the future can be used to transport molecular “loads”.
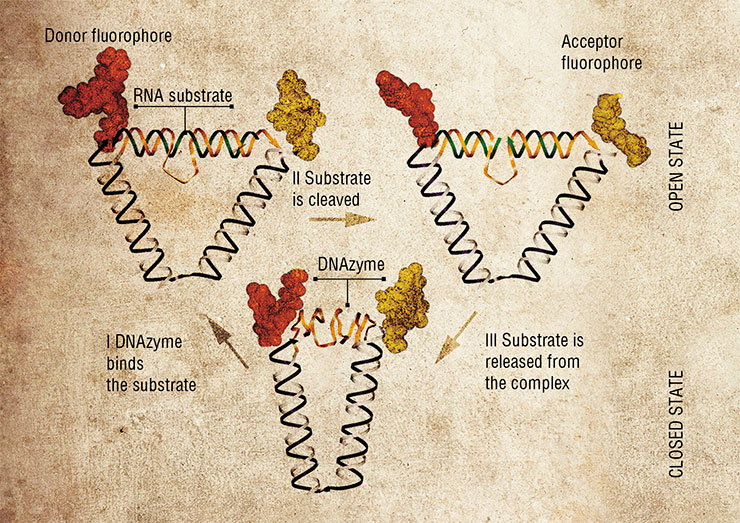
Molecules of nucleic acids hold promise for design of many other devices and compounds. Nanoswitches exploit the ability of double-helical nucleic acids to change their conformation depending on the environment, for instance, upon binding specific ligand compounds. If this sensitive fragment joins two elements of a nanoconstruct, adding the ligand reshapes the construct. In this way, for one, the fluorescence of a nanoobject can be changed.
The list of nanodevices cannot be complete without mentioning aptamers, a unique artificially designed class of nucleic acids. Aptamer may be regarded as a three-dimensional “key”, whose structure specifically fits the “lock” of another molecule, the target. Their interaction result in stable and lasting supramolecular complexes. This allows even minute amounts of the target compound to be detected. Aptamers are an indispensable part of the bionanoconstruction kit, which are used to design various supramolecular devices, including biosensors.
Diagnosis: mutation
While some achievements of bionanotechnology can still be regarded only as prototypes of not-so-near future devices (who can find a decent job for a nanomotor today?), others are immediately developed into applications of great importance, such as medical diagnostics. A good example is given by nucleic acid-based diagnostic sensors, many of them developed in the past 15 years at SB RAS Institute of Chemical Biology and Fundamental Medicine (ICBFM), Novosibirsk. These constructs are designed to recognize certain DNA sequences with high accuracy, due to specific binding of nanoprobes with a DNA target.
The diagnostic systems offered by the Novosibirsk biochemists consist of sets of short synthetic DNA fragments. These probes can bind the tested DNA forming so-called tandem complexes.
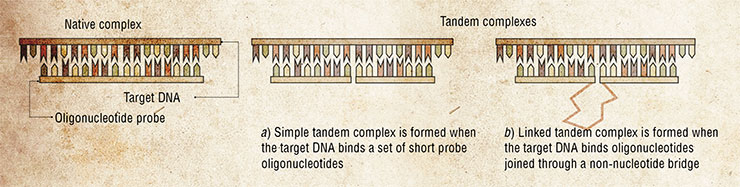
Years of basic research of these complexes culminated in development of test systems capable of revealing point mutations in DNA. Mutations of this kind often cause grave hereditary diseases and determine the pathogenicity of different strains of bacteria and viruses.
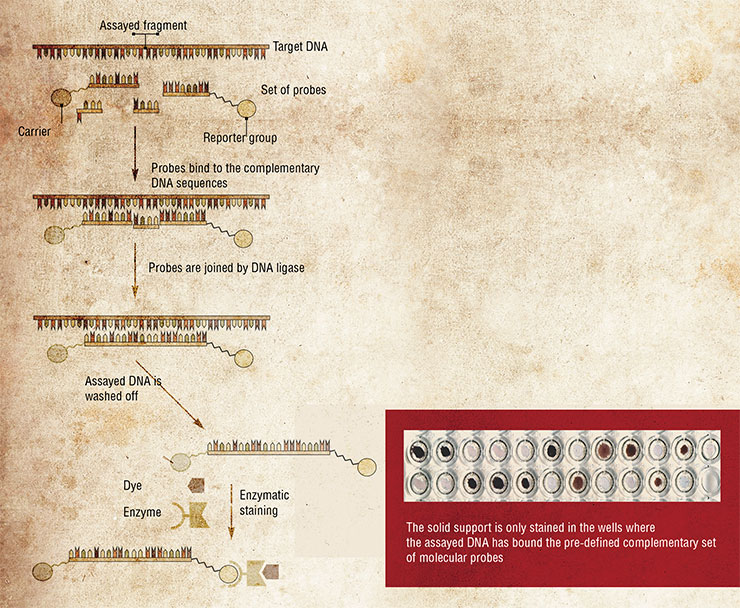
In addition, these test systems can identify other localized deviations in DNA sequences, such as loss of a DNA segment, replacement of several nucleotides, etc.
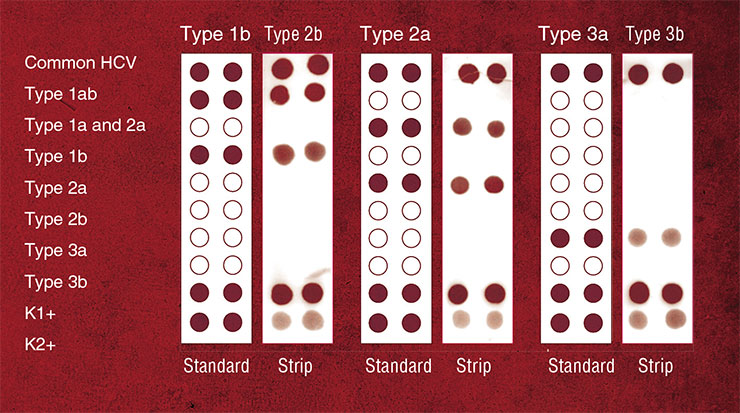
Another actively pursued direction of medical diagnostics making use of material developed with bionanotechology tools is design of new types of nanoprobes for state-of-the-art methods of quantitative DNA analysis, such as real-time PCR. This technique is used to simultaneously detect the target DNA and quantify the number of its molecules in the sample.
Drug delivery
One of the most important problems in clinical practice is that of addressed delivery of biologically active macromolecules, such as “therapeutic genes”, to their target cells. Any solution of this problem must provide both protection of the drugs during their transport and the means to concentrate them in the required cells.
Drugs based on nucleic acids face a major obstacle of low efficiency of their entry into cells, or transfection. The underlying reason is that mammalian cells possess several mechanisms restricting the uptake of foreign DNA and RNA molecules, a potentially harmful genetic material of viruses, bacteria, etc.
Besides, the cells have difficulties taking up free (“naked”, in the professional jargon) nucleic acids due to the existence of an electrostatic barrier. The cell membrane and sugar-phosphate backbone of nucleic acids are both negatively charged, which causes electrostatic repulsion between them.
The transport of long nucleic acids into cells is additionally complicated by their relatively large size, rigid three-dimensional structure and low mobility in biological fluids and cell cytoplasm.
The methods of transfection have been perfected for over three decades now. The most advanced ones engage complicated constructs based on nucleic acid nanocomplexes and their conjugates with organic ligands and nanoparticles.
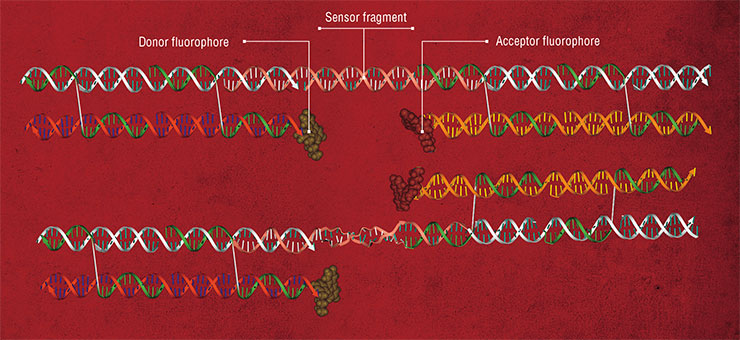
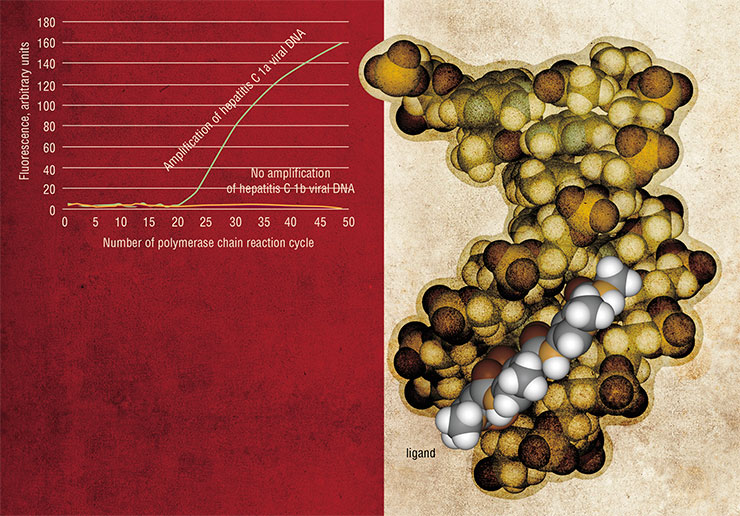
PICTURESQUE DIFFERENCES Oligonucleotide probes form a base of a new accurate and highly efficient assay called real-time PCR, which can not only detect the presence of a DNA target but also precisely quantify the number of its molecules. In a regular polymerase chain reaction (PCR), the target DNA presents a template for copying by DNA polymerase, and these copies serve as templates in the following rounds of DNA synthesis. In the end, the minute amounts of target DNA are amplified to a great extent. The unique feature of real-time PCR technique is the ability to follow the amount of DNA as it is built up in the reaction. The most promising variant of the method is based on the so-called TaqMan probes, oligonucleotides with a fluorescent label at one end and a fluorescence quencher at another end. The quencher absorbs the emission from the fluorophore and makes it invisible for a photodetector. Moving DNA polymerase can degrade probes that have annealed to the template in its way. As the TaqMan probes anneal to the complementary parts of the DNA template, they are destroyed by the polymerase, the fluorophore is separated from the quencher, and detectable fluorescence eventually increases. One disadvantage of this method is its low ability to discriminate between DNA targets that differ from each other by only one nucleotide. A solution proposed in the SB RAS ICBFM Laboratory of Medicinal Chemistry is to use the probes consisting of short oligonucleotides conjugated with minor-groove ligands, synthetic molecules that tightly bind to the DNA double helixSome RNA molecules occurring in nature have the ability to form compact molecular complexes. One of them is 29, a short (117 nucleotides) RNA of the bacteriophage of the same name, which participates in the packing of the phage DNA genome into the protein coating. This RNA was used as a starting point to design “packing RNA” (pRNA). Forming hydrogen bonds between its separate domains, pRNA can self-assemble in dimers, trimers and hexamers up to 10–30 nm in size (Shu, Huang et al., 2003; Khaled, Guo, Li et al., 2005).
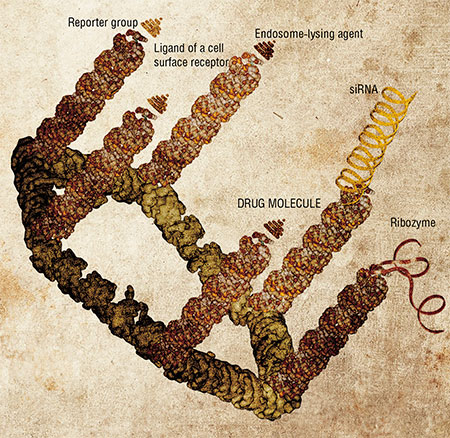 Such pRNA complexes serve as the packing scaffold for therapeutic nucleic acid molecules. They can also be functionalized (another word from the biospeak) by adding various moieties with the aim to specify the address of delivery of the construct and impart other abilities to it. These can be reporter groups, which allow one to estimate the transfection efficiency; gene-targeted molecules (ribozymes or small interfering RNA, siRNA), which disrupt the execution of pre-defined genetic programs in the cell; or ligands for cell surface receptors and membrane proteins, which define the “address” for the capture of the nanoconstructs by their target cells.
Such pRNA complexes serve as the packing scaffold for therapeutic nucleic acid molecules. They can also be functionalized (another word from the biospeak) by adding various moieties with the aim to specify the address of delivery of the construct and impart other abilities to it. These can be reporter groups, which allow one to estimate the transfection efficiency; gene-targeted molecules (ribozymes or small interfering RNA, siRNA), which disrupt the execution of pre-defined genetic programs in the cell; or ligands for cell surface receptors and membrane proteins, which define the “address” for the capture of the nanoconstructs by their target cells.
An example of an addressing group is a well-known compound folate (folic acid, vitamin B9). The surface of normal differentiated cells usually lacks receptors to it, but a lot of the receptor molecules are exposed on the outer membrane of tumor cells of various origins. Therefore, a folate moiety attached to a nanoconstruct will secure its preferential delivery to the tumor cells.
Experiments on targeting these pRNA complexes with a biologically active oligonucleotide (ribozyme or siRNA) demonstrated that they efficiently penetrate into the tumor cells that carry folate receptors and suppress the work of the target genes.
Since a pRNA molecule can form hexameric complexes, its functionality can be expanded by adding up to six different moieties. As a result, the selectivity and efficiency of the therapeutic nanoconstruct should be significantly enhanced.
Self-assembly of pRNA-based nanoparticles is a controllable process, thus allowing the researchers to adjust their size by manipulating the structural domains. pRNA dimers and trimers form 20–40-nm particles. Such structures are large enough to prevent their fast elimination from the circulating blood but do not reach the critical size (> 100 nm), which hampers the entry of the complexes into cells.
Cholesterol assistant
An alternative way around the problem of delivery of therapeutic nucleic acids into cells is to raise the efficiency of their natural transport through the cell membrane. This approach also requires formation of various supramolecular structures.
In particular, long oligonucleotide nanocomplexes developed in SB RAS ICBFM better penetrate into the cells from various tissues due to their higher affinity for the cell phospholipid membranes. These nanoconstructs are concatemeric complexes, long double-stranded DNA molecules with overlapping complementary nucleotide sequences. One of the strands (the address molecule) is the biologically active molecule, subject to delivery into the cell, whereas the other strand is the transporter molecule (Gusachenko et al., 2008).
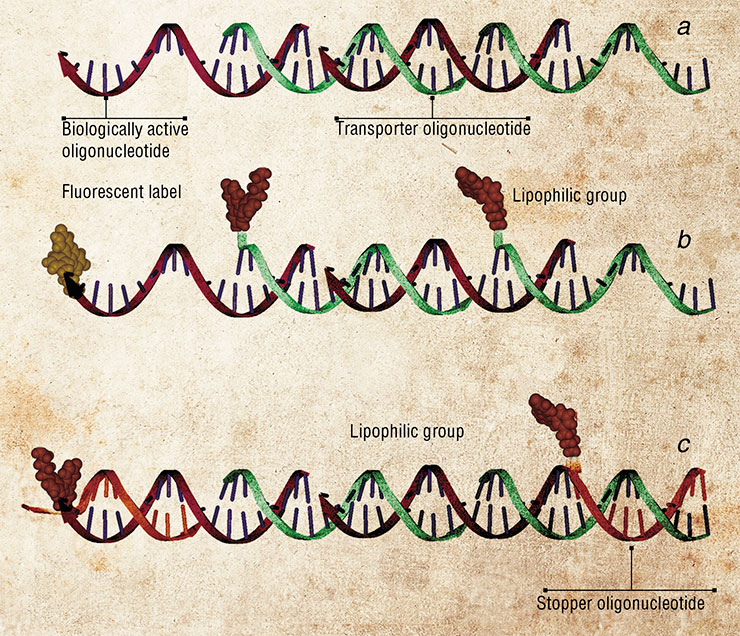
The functional group in the transporter oligonucleotides is a lipophilic cholesterol moiety, which facilitates the uptake of the construct due to its high affinity for the phospholipid cell membrane. Worthy of note, the functional moieties are linked to the transporter leaving the therapeutic molecule intact to spare its high biological activity.
The problem of addressed delivery of drugs to their target organs and cells is central to modern medicine. Bionanotechnologists employ nucleic acid to create the scaffolds capable of carrying several functional groups, which allow the constructs to successfully cross the barriers on their way and transport the bioactive molecules to their targetsThis delivery system has been tested in the experiments on the transport of antisense oligonucleotides that switch off the gene encoding the green fluorescence protein. In the cells that make this protein continuously and fluoresce green, this nanocomplex caused the fluorescence to drop by 30 %, indicating that the antisense oligonucleotide had entered the cells and specifically suppressed the action of the gene for the green fluorescent protein.
Nowadays, most bionanotechnology research is conducted as interdisciplinary projects. Success in this field critically depends on the close collaboration of research groups from different areas of science.
This organization model is nicely illustrated by the joint work of Novosibirsk biochemists and their colleagues from the SB RAS Institute of Semiconductor Physics who specialize in production of unique micro- and nanoporous silicon-based membranes. These materials can be advanced for applications in state-of-the-art biosensor devices, DNA diagnostic kits and for ultra-selective isolation of target cells.
Cooperation and integration of researchers and projects is key to success in modern bionanotechnological studies. These principles are successfully implemented in the Siberian Branch of RASAnother interdisciplinary project is concerned with the development of micro- and nanofluidic devices for amplification and analysis of nucleic acids, carried out together with the researchers from the SB RAS Institute of Catalysis.
Besides all that, SB RAS institutes working in the fields of chemistry, physics, and even petrography, are lining up to offer materials with possible bionanotechnological applications: nanopowders (nanospheres, nanotubes, quantum dots) and nanoporous materials.
The possibilities that SB RAS integration projects provide for such sharing of materials and technologies open the way for making Siberian high-tech “intelligent” biosensors and smart drugs that will shape medicine and biotechnology of the future.
References
Aldaye F. A., Palmer A. L., Sleiman H. F. Assembling Materials with DNA as the Guide // Science. 2008. V. 321. P. 1795–1799.
Chen Y., Wang M., Mao C. An autonomous DNA nanomotor powered by a DNA enzyme // Angew. Chem. Int .Ed. Engl. 2004. V. 43. P. 3554–3557.
Gusachenko (Simonova) O. N., Pyshnyi D. V., Vlassov V. V., Zenkova M. A. Modified concatemeric oligonucleotide complexes: new system for efficient oligonucleotide transfer into mammalian cells // Hum. Gene. Ther. 2008. V. 19. P. 532–546.
Khaled A., Guo S., Li F., Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology // Nano Lett. 2005. V. 5. P. 1797–1808.
Shu D., Huang L. P., Hoeprich S., Guo P. Construction of phi29 DNA-packaging RNA monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices // J. Nanosci. Nanotechnol. 2003. V. 3. P. 295–302.
Simmel F. C., Dittmer W. U. DNA Nanodevices // Small. 2005. V. 1, N. 3. P. 284–299.
Tian Y., He Y., Chen Y. et al. DNAzyme that walks processively and autonomously along a one-dimensional track // Angew. Chem. Int. Ed. Engl. 2005. V. 44. P. 4355–4358.