Saved by the Cell: How Cells Repair DNA
All information kept and handed down by the living cells is carried by the DNA molecules — encoded genetic “texts”. A cell thoroughly looks after its treasures: DNA is the only molecule that the cell repairs when it is damaged, while all other molecules are synthesized anew
Despite the key role that the DNA strand plays in cell work, it is extremely vulnerable and is very easy to damage. Even ordinary sunlight is a serious threat for DNA. Usually it happens like this: a light quantum, a photon, having collided with DNA can transmit its energy to one of the DNA’s structural elements, the nucleobase, which acquires an excited state. Then everything depends on which particular base was affected. If it is adenine or guanine, then excitation energy quickly turns into thermal energy, and the DNA structure does not change. If the portion of the additional energy is given to thymine or cytosine, then the consequences can be rather serious.
For example, two adjacent thymines can form a new structure, the thyme dimer: a molecule, in which four atoms of carbon situated on the corners of a square are connected by covalent bonds. If we picture DNA as a zipper, the dimer reminds us of two adjacent teeth stuck together that interfere with the zipper: the damaged DNA strand cannot be copied so as to create an “instruction” for making a necessary protein.
Another quite harsh blow for any organism is radiation that is several times higher than the background level necessary for all living organisms. Ionizing radiation leads to various DNA lesions, including double strand breaks, which are the most dangerous lesions for the cell due to an especially complex repair. The principle serves as the basis for radiotherapy when the ionizing radiation is used to destroy cancer cells. Some anticancer drugs used for cancer treatment also employ this mechanism. Just one not repaired double strand break in DNA can kill the cell.
The DNA damage can be triggered by anthropogenic pollution of environment with combustion products and tobacco smoke… However, the most numerous and at the same time the easiest to repair are lesions caused by the agents appearing in the process of normal life of the cell itself: breathing, oxidation of lipids, and inflammation. Today more than one hundred types of such oxidative lesions of DNA are known.
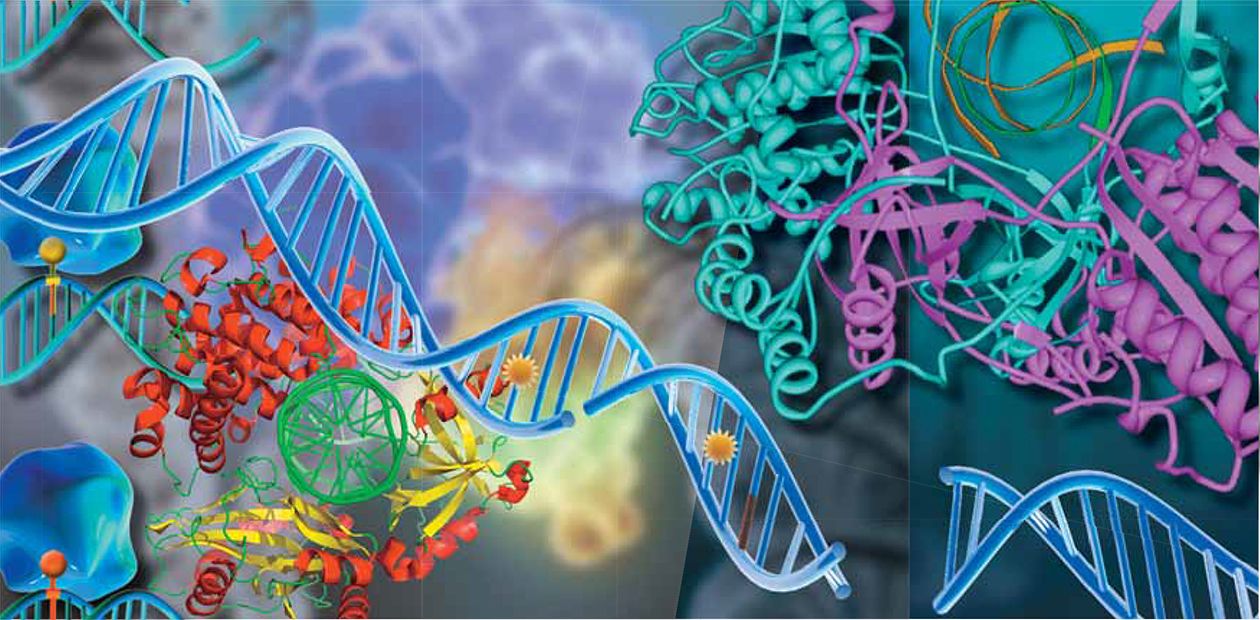
Evidently, it is not easy to cope with such various lesions. In the process of evolution, several elaborated, partially overlapping systems have appeared that are able to repair most (but not all) changes in the genetic cell “texts”. As a rule, the systems are ensembles of several dozen various proteins, and they specialize in certain types of DNA repair.
The protein “machines” divide the functions between themselves as follows. The nucleotide excision repair provides for excising a large class of various bulky lesions which distort the structure of the DNA duplex and, as a rule, hinder the synthesis of DNA copies, that is, inhibit obtaining copies-instructions for the subsequent protein synthesis.
The base excision repair specializes in “fast” repair of small DNA lesions not accompanied by significant distortion of double helix. The lesions do not necessarily interfere with the process of copying the DNA information, but they lead to mistakes in the very genetic “texts”. There are two more protein groups taking part in the processes; they are named “homologous recombination” and “non-homologous end-joining” , and are responsible for the heavy maintenance, joining the DNA strands with both strands broken, while employing different mechanisms for that.
Short and long patches
Let’s see in more detail how some repair machines work. Despite many differences in the type of the damaged DNA repair and in the “executors”, all repair processes in cells share some certain features.
First of all, it is necessary to find the lesion, “clear” the damaged part of DNA (it may be a small part, just one nucleotide in size, or a more lengthy piece of several dozen nucleotides), then to patch the lesion and finish it so that there is no trace of the patch. Another common feature is that the damaged parts are not repaired but replaced with new ones. The repair process uses the same ready-to-use units that form the DNA strand in the replication process. Nevertheless, the work of these machines differs in details.
During the base excision repair (BER), the DNA repair is carried out by a sort of a conveyor consisting of separate proteins. The damaged DNA is brought to the beginning of the conveyor and comes out of it completely repaired, that is, it is dispatched from one protein to another like a “baton”. Certainly, it is not DNA itself that moves. It is more like every protein comes around and begins working just as the previous participant of the process completes its task.
In the very beginning of the conveyor there are several enzymes, DNA glycosylases, which find the damaged base and excise it*. Some of them find and excise several different types of damaged bases, that is, can “bite off” from the DNA strand differently damaged “beads” without cutting the strand itself. Others “notice” only some definite type of damage.
Then an enzyme called apurinic/apyrimidinic endonuclease (APE1) incises the strand so that another enzyme, DNA polymerase, could attach a new undamaged “bead” together with a piece of strand and at the same time could “trim” it by excising the scraps of the damaged bead (i. e., the deoxyribosophosphate residue at the other side of the nick). As the second strand of the “necklace” is intact, the DNA polymerase picks out of stock not any ready-to-use unit, but the one complementary to the bead opposite the lesion. DNA ligase joins the strand ends, and now it is impossible to guess that the “necklace” was repaired.
The described mechanism is the easiest way of the base excision repair. It is called “short-patch”, for it replaces only one “bead”. It is the predominant way the BER system repairs lesions. The process also employs auxiliary proteins which do not perform separate operations but help other proteins to do their best. Such accessory proteins include, for example, the protein with the enigmatic name XRCC1, which provides resistance against X-rays: cells lacking the protein are more sensitive to the ionizing radiation effects, and the DNA repair is poor in them.
As you can see, BER is an economical and well-arranged way of repair. However, not always everything goes so smoothly. If a piece of the strand left from the damaged bead which was supposed to be excised by DNA polymerase b was also damaged, it is not excised, and the cell switches to another way of repair.
In doing so, the cell can use help of other protein machines, for example, of those DNA polymerases that are usually employed not in repair but in the assembling of new DNA strands. Such DNA polymerases along with their accessory protein PCNA work faster and make fewer mistakes when picking the type of a ready-to-use bead. The old strand is driven out of the “necklace” and forms a dangling “tail”. This is where yet another protein is needed to excise the “tail”. An enzyme called flap endonuclease (FEN1) also belongs to the replication machine and does its best when interacting with PCNA.
The given mechanism, which was discovered a bit later, was called a “long patch”, because it replaces the piece of DNA consisting of not one but several nucleotides. To mend the end break in this case, one more “replicative” enzyme is used, DNA ligase. The DNA with the breaks due to ionizing radiation is repaired in the similar way. As a rule, the process requires the participation of a further enzyme, polynucleotide kinase, whose role is to create a certain structure of the strand ends which is convenient for the work of the enzymes DNA polymerases and DNA ligases.
Too few or too many is too bad
When the nucleotide excision repair (NER) is employed, a group of proteins comes to the identified DNA lesion, unwinds DNA near the lesion, and excises it. The lesion is excised along with a piece of normal DNA.
It is considered that lesion recognition is the responsibility of the protein XPC-HR23B. (Many of the proteins in the group are indicated with the initial XP, connected with xeroderma pigmentosum, meaning higher sensitivity to sun light due to the absence of one of the NER proteins.)
Proteins of the repair machine take their places in strict order, and “landing” of each protein facilitates the functioning of the next participant of the process. The work of excising a piece of DNA of 30 nucleotide residues is well organized. Helicases with their accessory proteins separate the DNA strands, and the replication protein A (RPA) covers and holds the undamaged strand. The nucleases with their accessory proteins make two incisions of the strand on both sides of the lesion. The incisions are made so that later the ends of the strand will be convenient to restore the damaged part with the replication machine. The cut part can be removed, and the job is half-done in terms of preparatory work for restoring the missing part.
Now the proteins of the replication group can be helpful: the DNA polymerases with their assistant proteins build up the missing part of DNA link by link using information of the undamaged strand, and the DNA ligase seals the strand break. The job is done, but this way of repair is ambiguous.
There are two kinds of protein ensembles, which are different in composition and specialization. One constantly traces or scans the DNA strands and repairs them as needed. The other is a “team of swift response”. It enters the scene when the enzyme RNA polymerase (also, together with its accessory proteins — it is impossible to accomplish anything worthy alone, is it not?) begins to compose on the DNA strand another nucleic acid, the so-called messenger RNA, which serves as the instruction for synthesis of a needed protein**. If there is a bulky lesion in DNA not allowing RNA polymerase to decode information and to write it in the instruction, the process stops: this is where the “team of swift response” is invited to repair the lesion in the way described above.
We will not touch upon the work of two more systems specialized in the repair of double-strand breaks in DNA — let us leave it for another time.
To ensure the right and timely DNA repair, it is important for all participating proteins not only to be present in the cell, but to be present in the right amount: both lack and abundance of the proteins are dangerous. What threat for humans do the defects in the DNA repair systems pose? Such diseases are serious but rare (the most well-known is the above mentioned xeroderma pigmentosum).
Human diseases unambiguously linked to the functional inactivity of a protein in the BER system are not known. It can be explained by the great significance of the described way of repair, resulting in the lack of viability of organisms with such defects.
At the same time many pathological states are characterized by a higher or lower number of proteins. For some types of cancer cells an increased amount of DNA polymerase b and APE1 is typical. Why having a lot of repair enzymes in the cell should be a problem? The matter is that a dramatic change in the activity (amount) of, for example, DNA polymerase b can involve it in doing somebody else’s job: it replaces the DNA polymerases that function more precisely, which results in the instability of genetic information.
Labeled proteins
To study protein complexes dealing with replication and DNA repair systems as well as to find new participants of these processes, scientists use the method of the so-called photoaffinity protein modification developed at the Institute of Chemical Biology and Fundamental Medicine, SB RAS. “Affinity” means “based on affinity, on relationship”. It means the ability of DNA to form specific complexes with proteins participating in certain reactions.
The scientists designed and synthesized the analogs of deoxynucleoside triphosphates (ready-to-use units used by DNA polymerases when synthesizing DNA) which display the following features: they carry additional chemical groups that are activated under ultraviolet light and can cross-link to the protein, forming a stable bonds. The photoreactive groups are attached to the base so as not to interfere with the work of DNA polymerases that can insert the photoreactive “beads” into DNA.
Now, if we expose the mixture of the photoreactive DNA and proteins to UVlight, the proteins which form a specific complex with DNA will bond with it strongly. In this way the protein obtains a “label” in the form of a part of DNA.
As a result, reserachers kill two birds with one stone. First, since DNA includes atoms of phosphorus,we can introduce a radioactive label into its composition (normally, the 32P phosphorus isotope), which gives us a chance to easily watch the marker in further analysis. The radioisotope labels are registered with high sensitivity, which allows us to work with minute amounts of proteins. Second, the label present on a protein testifies that the protein is able to interact with certain DNA structural peculiarities characteristic of one or another stage of the repair process.
Using the method, the reserachers of the Institute of Chemical Biology and Fundamental Medicine have revealed that, for example, out of thousands of proteins contained in a mouseous cell extract only six turned out to be radioactively labeled with photoreactive DNA, analogous in structure with the DNA intermediates of the BER process. Among them there are our “old acquaintances” — DNA polymerase b, flap endonuclease, apurinic/apyrimidinic endonuclease, all of which are well-known for their participation in this way of repair.
Modern fine technologies can help in identifying of the protein marked with the affinity modification method. As DNA-labels, one can use specific DNA molecules containing AP-site, sort of a “hole” in the place of one nucleobase, which appear as interim compounds during excision repair. Sugar deoxyribose in the AP-site can form chemical bonds with certain amino acids of the protein, the bonds becoming stable when reduced by borane.
To separate such a complex, a very fine and clever method is used. First, you add biotin to DNA (vitamin H or B7), which is able to bond strongly with the chicken egg protein avidine or bacterial protein streptavidine. If streptavidin is attached to some surface and then the surface is covered with the solution of DNA with biotin, the latter will stay there without yielding to water solutions. In this manner you can “anchor” DNA with an attached protein, too. Streptavidin is secured on small paramagnetic balls, which are held by a magnet when washed.
The product left on the balls is taken away and separated from the admixtures by electrophoresis in polyacrylamide gel. The piece of the gel containing the protein with attached DNA is excised and “cut” in certain places into pieces-peptides right in the gel with the help of enzymes such as trypsin.
The mass of the formed peptides is determined by a very sensitive method of mass-spectrometry, MALDI-TOF-MS. The analysis gives a “peptide map” of the protein. Then with the help of such software as Mascot or Profound identification of the protein is performed, for which the mass of peptides in the given sample is compared with the theoretically calculated masses for all proteins in the database. Upon processing a great body of data, the program chooses the most probable candidates. Peptides with identical masses can be encountered in different proteins, but the combination of peptides of a certain mass in the sample allows to determine the nature of the analyzed protein to a high degree of certainty
The nature of one more protein was determined using other methods, including specific antibodies. The protein is the enzyme poly (ADP-riboso) polymerase 1 (PARP1). It can find breaks in DNA strands, forming dimers (pairs), and, as a response, it synthesizes a branched negatively charged polymer (poly(ADP)ribose) strongly bonded with some nuclear proteins. The most amazing thing is that one molecule PARP1 synthesizes this polymer on the second member of the pair. This is how the intracellular signal appears: “Attention! A break in DNA has been found. Help needed”.
As a result, a cascade of events is triggered in the cell, which in the end leads to the lesion repair. If lesions are numerous, “hyperactivation” comes in, that is, making a great amount of the “signal”. It exhausts the energy stock of the cell, and another “signal” is formed: the capacity for DNA repair is exhausted, and the cell is to be destroyed. In response to the signal, programs of the cell self-destruction come into effect, for the cells with DNA lesions can be parental for tumor cells.
Two more of the marked proteins keep their “incognito”, but the work to establish their identity goes on. Today scientists have learned to determine the sequence promptly and precisely and, basically, “have read” all genetic “texts” written in human DNA; however the functions of many proteins that can be synthesized by the cells have not been established yet. It is a vast field for the researchers of today and tomorrow.
*SCIENCE First Hand, 2006, № 7 (12), pp. 18—29
**SCIENCE First Hand, 2006, № 7 (12), pp. 40—47.