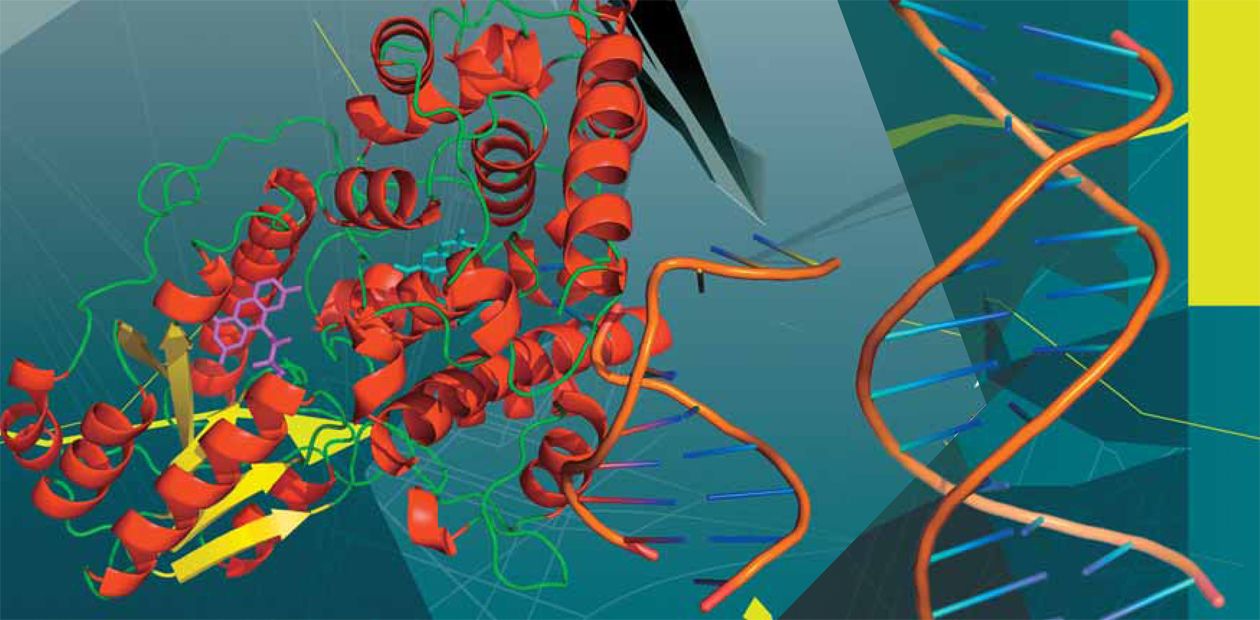
Sentinels of Genome
DNA, the carrier of our hereditary information, is attacked every single moment by various adverse factors. It is not only the notorious "bad ecology" - chemical pollution of environment, radiation, or ultraviolet. In the cells, numerous substances involved in cell metabolism can damage DNA, and even ubiquitous water can pose a hazard. Damages in DNA structure are fraught with mutations and even cell death. The result is hereditary diseases, cancer, and aging… Luckily, a squadron of intricate repair systems backing up the work of each other guards our genome.
This story began as in the Persian fairy tale poem Eight Gardens of Paradise by Amir Khusraw Dahlawi, dating back to the 14th century. Once upon a time there were three brothers, princes, and they lived in the Island of Ceylon, named Serendip at that time, which is the Sanskrit name for Sri Lanka. The special thing about them was that when they were looking for something, they accidentally discovered something completely unrelated but even more necessary. Almost five centuries later, H. Walpole, an English writer, coined the word serendipity. This word is among the ten English words most difficult for translation; however, its essence is clear from the legend – this is the ability to make most unexpected fortunate discoveries by accident. No wonder that this word is especially frequently used in the scientific world.
A DNA molecule is a helix formed of two strands. The strands are composed of successively bound nucleotides, nitrogen bases attached to sugar (deoxyribose) residue. This gives an integrated sugar–phosphate backbone with attached bases of four different types–adenine (A), guanine (G), thymine (T), and cytosine (C). The bases from two strands are linked to each other via hydrogen bonds following the principle of complementarity (A with T and C with G), making the overall construct stable. Adverse factors of various natures can damage individual bases, nucleotides, and the sugar–phosphate backboneThus, everything happened in a serendipitous manner… In 1946, A. Kelner from the famous Laboratory of Molecular Biology in Cold Spring Harbor was involved in important work – he was searching for new antibiotics. This was just after the war; the success of penicillin was fresh in everybody’s mind. Experimenting with the bacteria streptomycetes, Kelner decided to find out whether it was possible to make them produce a larger quantity of the target product by exposure to some stress factors. He used ultraviolet as a stress factor. The trouble was that he failed to choose a necessary irradiation dose that would provide for survival of bacteria. The data on survival rates of streptomycetes estimated according to the growth of their colonies “jumped” depending on the conditions under which they were kept after irradiation.
Luckily, being very punctual, Kelner meticulously recorded every detail of his experiments. It appeared that immediately after irradiation the survival of streptomycetes was low. The situation did not improve much if the cells were kept in darkness for some time. However, in the light, the survival rate gradually increased, and the more light, the quicker was the increase!
It took Kelner three years to countercheck all results. Meanwhile, R. Dulbecco, a future Nobel laureate and at that time a young researcher with the team of S. Luria, also a future Nobel laureate, studied the inactivation of bacteriophages (viruses that infect bacteria) by ultraviolet. Dulbecco came to the same conclusions as Kelner.
An amazing coincidence: when Kelner prepared the paper describing his results, he sent it to Luria for reviewing. To their credit, Dulbecco and Luria did not hold out the paper of their competitor; on the contrary, they shared their achievements with Kelner. Both papers appeared simultaneously. By and large, it was already known at the time that ultraviolet damaged the cell’s genetic material, whereas the results in question suggested that visible light enhanced the restoration of genetic material. This process was named photoreactivation.
This was how, very casually and by chance, DNA repair was discovered – and this process is not less important than the other major molecular genetic processes of replication, transcription, and translation of genetic material. The story of the princes of Serendip took place once again…
What’s the big idea?
As is known, DNA consists of nucleobases of four different types and a sugar-phosphate backbone, forming a continuous chain with attached bases. Both the bases and the backbone can be damaged. Totally, about one hundred of various possible lesions can be produced by different mechanisms.
For example, deamination (removal of an amino group, NH2) converts cytosine, a common base for DNA, into uracil, normally present only in RNA. DNA bases can be oxidized. Other carbon-containing groups can readily bind to certain positions in the bases – this process is referred to as alkylation. Ultraviolet can induce a crosslinking of two neighboring thymines, forming a dimer. Very frequently, a base can be lost from DNA, thereby producing AP sites. As for ionizing radiation, it causes single- or double-strand breaks in DNA.
The DNA lesions, especially when they are numerous, can cause not only mutations, but also cell death. Note here that the DNA damage and mutation are not the same things. Replication – synthesis of a new DNA strand on the “old” strand used as a template – is necessary for a damage to become a mutation. Usually, replication follows the principle of complementarity. However, if a template contains a damaged element, a wrong base may be incorporated into the daughter strand opposite to the lesion.
In particular, just anything can be put at the position opposite to an AP site, because the template contains no instructing base. This gives rise to a mutation, i.e., an alteration in the DNA sequence as compared with the initial template. Mutations in germline cells are inherited by the next generation, while they can lead to cancer in the somatic cells. Many researchers believe now that the most incurable “disease”, aging, is also tightly connected with accumulation of mutations in the cells of our body.
DNA REPAIR
Currently six different DNA repair mechanisms are known; they work independently but sometimes can interact. In addition, one repair system does not fix damaged DNA but assists the cell to survive.DIRECT REVERSAL.
This process requires one specific protein – the enzyme that corrects only one specific lesion. Among them are the suicide enzymes, capable of catalyzing only a single reaction, i. e., correcting only one error. When they do it, their structure changes, making them unable to work further.BASE EXCISION REPAIR.
Small damaged bases that do not distort considerably the DNA structure are repaired by this pathway. Initially such base is recognized by one of DNA glycosylases, which excises it from DNA. There are over ten such enzymes, each excising damaged bases of a particular kind. This excision leaves an empty space (AP site) in DNA, which is eventually filled with the proper base.NUCLEOTIDE EXCISION REPAIR.
This mechanism, one of the most intricate repair mechanisms, is utilized in the case of the damages considerably distorting DNA structure. It comprises two branches. In the first, a damaged base can be located in any place of the genome; in the second, a damaged base can be found only in the comparatively few DNA regions that serve as a template for RNA synthesis (as is known, the larger part of our DNA is noncoding). Over twenty various proteins are involved in this process.In the global genome repair, the main role is played by the proteins whose names begin with the letters XP (abbreviated from the disease caused by their malfunction, xeroderma pigmentosum). The damage is recognized, and the DNA double helix is separated into individual strands. Then a region 27—29 nucleotides long is excised from such an “eyehole” to synthesize a correct missing fragment in its place.
Transcription-coupled repair is somewhat simpler; it takes place during synthesis of mRNA, which further serves as a template for protein synthesis. In this case, the damage is recognized by RNA polymerase, which is merely halted at the trouble site during DNA synthesis.
MISMATCH REPAIR.
A mismatch, a pair of noncomplementary nucleotides, is usually formed when the enzyme DNA polymerase makes a mistake in copying DNA. Since both nucleotides of the pair are undamaged, it is necessary for a proper repair to distinguish which of them was in the template strand and, therefore, should not be excised.This task is performed by a set of proteins that search for the nearest break in DNA strands – with a high likelihood, this will be the place where the DNA polymerase guilty of this error is located. Therefore, this particular strand was synthesized erroneously. The entire daughter strand up to the mismatch is destroyed to synthesize it anew.
NON-HOMOLOGOUS END-JOINING.
DNA helix can contain double-strand breaks. During repair, several nucleotides at the ends of the break are cut off and the strands are linked.However, the DNA fragment carrying the break is lost. Therefore, this process is mainly characteristic of higher organisms,whose genome contains predominantly nonfunctional DNA, making low the risk of losing the coding DNA. This type of repair is also necessary in the rearrangement of the genes encoding immunoglobulins, which is aimed at creation of new sequences.
HOMOLOGOUS RECOMBINATION.
This rather complex process provides for errorless “mending” of double-strand breaks. For this purpose, an undamaged copy of the same sequence is used, for example, the corresponding sequence from the other chromosome of the diploid chromosome set. It is difficult to describe this process in a few words, since it involves many enzymes and the DNA strands are shuffled hither and yon as cards in the hands of a skilled illusionist.TRANSLESION SYNTHESIS
is not used for repair but rather for bypassing DNA lesions when the DNA polymerase that synthesizes a daughter strand has already encountered a damaged site and stopped. Then a regular DNA polymerase is replaced by another enzyme from the large family of translesion polymerases. Traditionally, these enzymes are designated with letters of the Greek alphabet; they are so numerous that it is still unclear which will run short first, the polymerases or the Greek letters.A specialized polymerase inserts one or two random nucleotides into the damaged site and gives place to the common polymerase. In this case, the damage is not removed, and a mutation can appear at this site. Nonetheless, the cell survives.
By the way, another source of errors in DNA is incorrect incorporation of nucleotides during regular replication. The enzymes named DNA polymerases make such mistakes at a frequency of 0.001—0.00001. Polymerases can sometimes correct their mistakes themselves, immediately cutting off the incorporated incorrect nucleotides, which further increases their robustness 100—1000-fold. However, even such preciseness cannot rule out tens or even hundreds of mistakes appearing in each our cell during its division.
All the errors introduced into a DNA sequence must be corrected. Therefore, during the evolution several systems generally referred to as “DNA repair systems” have emerged; they protect us from mutations caused by the damages of genetic material. It is practically impossible to describe these systems even briefly in a short paper: their detailed description in the Talmud of researches involved in the studies of DNA repair – DNA Repair and Mutagenesis (2006) by E. Friedberg and colleagues – comprises 1,500 pages of densely packed text. Nonetheless, it is quite possible to get an overview of how these systems work.
Physician, heal thyself!
In the case of a regular operation of the repair systems, our genome is quite stable. However, similarly to any complex system, they have their Achilles’ heels, where a failure leads to breakdown of the overall mechanism.
The genes coding for repair proteins are similar to all other genes in that they can be also affected by mutations. If such a mutation interferes with the normal function of the encoded protein, the corresponding repair pathway is switched off. Then the organism acquires the so-called mutator phenotype, providing for a much easier emergence of mutations with all the ensuing adverse consequences. Defects in certain repair genes appear as hereditary diseases usually accompanied by an increased risk of cancer development; however, their manifestations can be quite diverse.
Perhaps the most known and best studied among such diseases is xeroderma pigmentosum. It has even played a part in fiction: fans of the thrillers by Dean Koontz can recollect that Christopher Snow, an amateur detective and the protagonist of the Moonlight Bay trilogy, suffers from this disease. Actually, this is not a single disease but rather eight of them with fairly similar symptoms. The majority of cases result from defects in the nucleotide excision repair system – deficiency in any of the seven XPA–XPG proteins, involved in its operation, or in the XPV protein, which is a DNA polymerase involved in the translesion synthesis.
Experts in repair issues like to scare the audiences at lectures or conferences with the photos of xeroderma pigmentosum patients. The main symptom of this disease is severe burns on the skin after a shortest exposure to the sun. The burn lesions are quickly replaced by moles; the skin becomes dry and fissured with eventual development of skin cancer at the age of seven or eight. The eyes are also affected; they react badly to sunlight. Less than half xeroderma pigmentosum patients reach the age of twenty. Luckily, this disease is quite rare, affecting only four individuals per a million of Europeans. However, the rate of xeroderma pigmentosum among Japanese is sixfold higher.
This disease also attracts the attention of scientists due to its versatile appearance. Damage in a single gene rarely causes different diseases; however, this is the case. The reason for this is that the same protein can act in different protein complexes with different functions in the cell. Consequently, one mutation can interfere with the ability of this protein to work in some complexes, whereas in other complexes it will retain its function.
In particular, some mutations in the gene encoding the repair protein XPD cause a typical xeroderma, while other mutations in the same gene result in a completely different disease, Cockayne syndrome, causing problems in growth and development during the first two years of the patient’s life. Consequently, such children develop dwarfism and cachexia, and their faces assume a characteristic “bird-like” shape with hollow eyes and protruding nose. The outer membranes of nervous fibers are destroyed, many neurons die, and calcium salts are deposited in the brain blood vessels, all this giving the impression that the nervous system in this syndrome is aging much quicker than the remaining body. The average lifespan of such patients is even shorter, about 12 years.
Another mutation in the same gene causes a third disease, trichothiodystrophy, when DNA repair is practically unaffected, whereas the synthesis of some keratin proteins is impaired. The result is brittle hair, scaly skin, and a practically complete loss of fatty tissue on the face.
Totally, there are several dozen inherited diseases resulting from compromised repair systems. Characteristic of these diseases, as could be expected from a mutator phenotype, are an increased risk of cancer development and, frequently, accelerated aging.
In particular, Werner’s syndrome, when a certain repair helicase does not work, faithfully reproduces the pattern of a regular aging with the only exception – the usual span of a human life is reduced to 30—50 years… Also, DNA repair deficiencies often lead to neurodegenerative diseases. Everybody knows that nervous cells do not regenerate; this is not completely correct, yet a typical neuron must serve a person throughout his or her life. During this time, its DNA accumulates numerous damages.
Cream with enzymes
Naturally, such powerful and specific protective tools as repair enzymes could not but attract the attention of physicians. It would be great to learn to use them for protecting our genome against damages! Theoretically, it is feasible but for one enormous challenge: nobody has devised a method how to make the protein molecules used as a drug penetrate into all cells of the body. Moreover, proteins in general poorly penetrate into cells; to achieve this, various tricks are used. For example, proteins are packaged into special membranous vesicles, liposomes, capable of fusing with the cell membranes and releasing their contents into the cells’ interior.
However, this method still does not allow the therapeutic enzyme to be delivered to all tissues, only to those easy to access, such as skin. Repair enzymes have already found medical application in this area: indeed, skin, frequently exposed to the sun, belongs to the organs most susceptible to genotoxic stress.
The US company AGI Dermatics produces creams with the liposomes containing photolyase and endonuclease V, two enzymes involved in the repair of cyclobutane dimers, which are formed in the DNA exposed to ultraviolet. A small pot of such cream costs about a hundred dollars and, according to the manufacturers, works much better than the usual protective creams. Perhaps this is true: unlike most cosmetic products, which have never been tested as strictly as drugs, the AGI Dermatics liposomes are now going through the last stage of clinical trials as a drug for prevention of skin cancer, including xeroderma pigmentosum cases.
Most likely, repair enzymes have a high potential as protectors for the barrier tissues (skin and mucosa) against genotoxic stress. Currently, DNA glycosylases, which correct DNA oxidative lesions, are being tested (at this stage, in animals) as a drug for prevention of lung cancer – lungs are the organ most massively attacked by oxygen radicals.
To suppress repair in the enemy!
Thus, it is good when our repair systems function properly. However, it is not good when these systems work properly in our enemies – pathogenic bacteria, viruses, and tumor cells. Is it possible to act on them by suppressing their repair? Such examples are already known.
However strange it may seem, it appeared easiest to use the repair systems as a target for drugs in cancer diseases. Many chemotherapeutics are known to act through damaging DNA of the cancer cell. Certainly, these damages are repaired, thereby decreasing the efficiency of drugs. Thus, the inhibition of repair is now regarded as a promising method for boosting the efficiency of chemotherapy. For example, benzylguanine, an inhibitor of the suicide enzyme MGMT (this enzyme “fixes” alkylated DNA bases, thereby interfering with the widely used antitumor agents, such as carmustine and temozolomide) is now at the last stage of clinical trials.
Repair inhibitors for controlling bacteria and viruses are work for the future. However, it is known that reproduction of some viruses, for example, poxviruses (including the smallpox virus), herpesviruses (herpes simplex and chickenpox viruses), and lentiviruses (HIV) requires the repair enzyme uracil-DNA glycosylase, either their own or borrowed from the host. Inhibitors of this enzyme are already being tested as antivirals.
As for bacterial pathogens, upon encountering them our macrophages try to kill the intruders by “oxidative burst”, releasing a tremendous amount of oxygen and nitrogen radicals, which damage bacterial DNA. Only a powerful repair system allows bacteria to survive in such collision: once we are able to suppress this system, it will be easier to the defenders of our body to cope with infection.
How are you, DNA?
It is clear that DNA damage can cause a lot of trouble. Is there any way to somehow estimate the “health” of DNA, i.e. to count the “errors” it contains? This is very important, in particular, for assessing the potential mutagenicity of environment, the effect of a new drug on a human body, and many other things.
One of such methods, referred to as a comet assay, was invented by Narendra Singh (National Institute of Aging, United States) in 1988. In the simplest variant, the number of double-strand breaks in DNA is counted. Individual cells (for example, blood leukocytes) are sealed in agarose blocks to destroy their membrane with special reagents and place them into electric field to conduct electrophoresis, familiar to all biochemists. The negatively charged DNA molecule should migrate to the anode, but as it is very long, it practically remains at the same place. However, if the DNA contains breaks, shorter fragments quickly “run away” followed by longer fragments, and so on. Thus, staining DNA with special dyes visualizes a characteristic pattern, a “comet tail”. The longer the tail, the more numerous are short fragments contained in DNA and the larger is the number of breaks and degree of damage.
Since the invention of the comet assay, its many variants have been devised for detecting damages of different types. For example, if electrophoresis is conducted under alkaline conditions, DNA double helix separates into individual strands. Consequently, we can see not only double-strand but also single-strand breaks and AP sites. If such a block is soaked beforehand in a solution containing a DNA glycosylase, it will be possible to determine how many damaged bases cleaved by this enzyme are present in the DNA.
In addition to improving comet assay variants, researchers are constantly searching for new objects. For example, several years ago German researchers proposed to use the comet assay for estimation of the state of natural aquatic bodies using freshwater sponges, which can be easily “disassembled” into individual cells. They even came to the Lake Baikal, collected sponges there, and proved that the Baikal water was healthy for genes.
A system utilizing the SOS response of bacteria provides for a simple and rapid estimation of environmental genotoxicity. It is known that some bacterial genes, including repair genes, are switched on only in the case of DNA damage. Each of such genes contains a specialized regulatory element at their beginning, a SOS box. If a reporter element (for example, the gene encoding green fluorescent protein, so popular among biochemists) is inserted under a SOS box control, then the modified bacterium under the conditions damaging DNA will give the corresponding colored signal. Thus, the fluorescing bacteria will indicate a hazard for human genes.
Several dozens of such biosensor constructs are currently available; some of them were even sent to space for studying the injurious effect of cosmic rays.
When the paper was almost completed, the author searched the PubMed database, which holds the summaries of almost all papers in the field of biomedicine ever published in the world.
A quick query for “DNA repair” gave 33 646 publications. Almost all “hot” directions in biology somehow overlap with the repair issue – there are papers on repair in stem cells and genotoxicity of nanoparticles… As is evident, interest in this issue is tremendous and will grow further, since safety of the personal genome is everybody’s concern.
And let us hope that the miraculous feature of the princes of Serendip, which underlay the discovery of DNA repair, will once again serve the good turn to the scientists working in this field.
References
Zharkov D. O. The Enigma of “Rusty” DNA // Science First Hand. – 2006. – No. 7. – P. 18—29.
Khodyreva S. N., Lavrik O. I. Saved by the Cell: How Cells Repair DNA // Science First Hand. – 2007. – No. 3. – P. 72—79.
The author and editorial board are grateful to Dr. Christa Baumstark-Khan (Institute of Aerospace Medicine, Germany) for the assistance in preparing the artwork.