RNA World: Yesterday and Now
It has been long considered that ribonucleic acid (RNA), the closest “relative” of the famous DNA, performs only utility functions in the body, being a mere messenger in most intricate cellular processes. Although it was recognized that RNA could play the leading role at early stages in the evolution of life, it seemed rather evident that this molecule had yielded precedence to more specialized molecules—catalysts and information carriers. However, the discovery of a multitude of regulatory RNAs, associated with the phenomenon of noncoding “dark genome”, literally turned upside down the concepts about the current “world of RNA” and gave an impulse to the search for and design of novel diagnostic tools and drugs
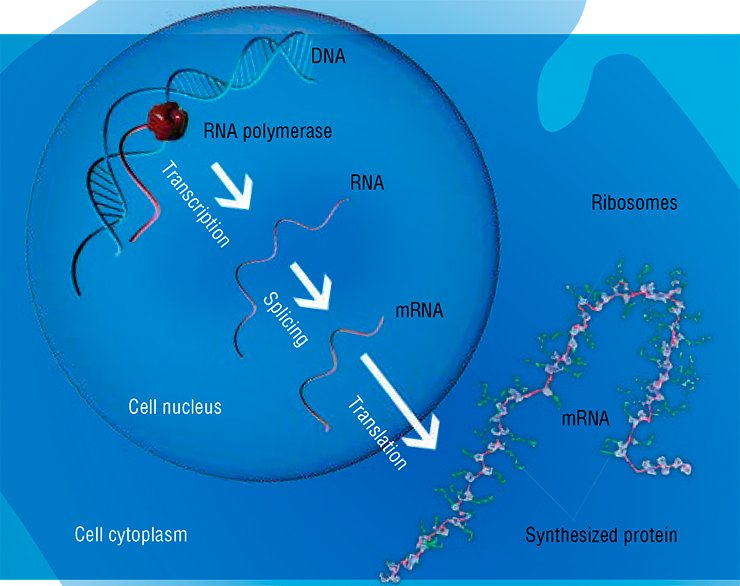
A classic view on the implementation of hereditary information in a living cell was formed as early as the mid-20th century. According to this view, all the hereditary information that determines the life of an organism is encoded as a sequence of nucleotides of a special biopolymer—deoxyribonucleic acid (DNA). This information is “reproduced” and transmitted via duplication of DNA molecules with the help of special proteins, enzymes.
When the DNA region containing information about the structure of a certain protein, referred to as gene, starts “working”, the special enzymes read the piece of “intermediate” information from its sequence in the form of ribonucleic acid (RNA) molecules. The nucleotide sequence of this RNA is the template “program” for operation of specialized molecular plants—ribosomes—which commence to synthesize the corresponding protein.
DNA is the central element in this scheme for the transformation of the genetic code into the final “usable” product. This concept was even considered “the central dogma of molecular biology,” stating that the information flow in a living cell goes exclusively in one direction—from DNA to RNA and further to protein: the cell is unable to synthesize DNA or RNA that would correspond to a particular protein. It is no wonder that a fascinating book by M. D. Frank-Kamenetsky, The Uppermost Molecule, published in 1983, was concerned with DNA.
An important role was also assigned to proteins as the major construction “bricks” and, first and foremost, catalysts able to mediate most diverse biochemical reactions and processes under the conditions of a living cell. As for RNA, it was represented as rather a “working” molecule, a consumable of protein synthesis. Certainly, both transfer RNAs, conveying amino acids to ribosomes, and RNAs within the ribosomal backbone were also known in addition to the messenger RNAs; however, the role of plain “Cinderella” was assigned to these molecules.
 The observations that made them wonder whether everything was so simply arranged in the genome had accumulated with time. For example, it was found that the hereditary information of certain viruses was encoded in their RNAs rather than DNA and they were able to synthesize DNA from an RNA “template” when inserting their genetic material into the host cell genome. However, despite certain contradictions, the views on the nature and roles of the most important biopolymers, included into all school textbooks, still remained hard and fast.
The observations that made them wonder whether everything was so simply arranged in the genome had accumulated with time. For example, it was found that the hereditary information of certain viruses was encoded in their RNAs rather than DNA and they were able to synthesize DNA from an RNA “template” when inserting their genetic material into the host cell genome. However, despite certain contradictions, the views on the nature and roles of the most important biopolymers, included into all school textbooks, still remained hard and fast.
Meanwhile, despite the difficulties of the experiments with RNA, the molecules of which are less stable than those of DNA and proteins, researchers tenaciously continued studying these “unfashionable” biopolymers. Thus, the classic scientific concepts at the end of the 20th century started to break down, crushed by new astounding discoveries.
Life started from RNA
Sequencing of the human genome as well as the genomes of other highly organized animals has demonstrated that the major part of DNA contains no genes coding for proteins. (By the way, comparison of the genomes of various mammals has shown that “the king of nature” in the set of genes very insignificantly differs from, say, the mouse.) They expected to find over 100 000 genes in the human genome with its length of over three billion nucleotide pairs, yet their real number is only one-third of that! Moreover, this set comprises not only all protein-coding genes, but also the genes of ribosomal and transfer RNAs. Together with all the regulatory sequences known at that time, the complete set of genes accounts for no more than 1.5% of the overall genome DNA. And what is the remaining part?
With a complete disregard of the incomprehensible, it was believed that such a noncoding DNA—the so-called dark genome—was “junk” that had accumulated during the evolution. Although it was found later that some information was still transcribed from the regions of “junk” DNA and that RNA was synthesized in a tremendous amount, this phenomenon was regarded as a “transcriptional noise.”
The most amazing discovery has been the fact that proteins are not monopolists in their ability to catalyze biological reactions. In particular, a set of small RNAs, discovered in the cell nucleus, together with proteins provide maturation of messenger RNAs (mRNAs). The fact is that a long RNA species is transcribed from a gene but should be later cut into fragments and than “stitched” together in a certain manner to get the necessary program. The correctness of this “stitching” is provided for by special RNAs.
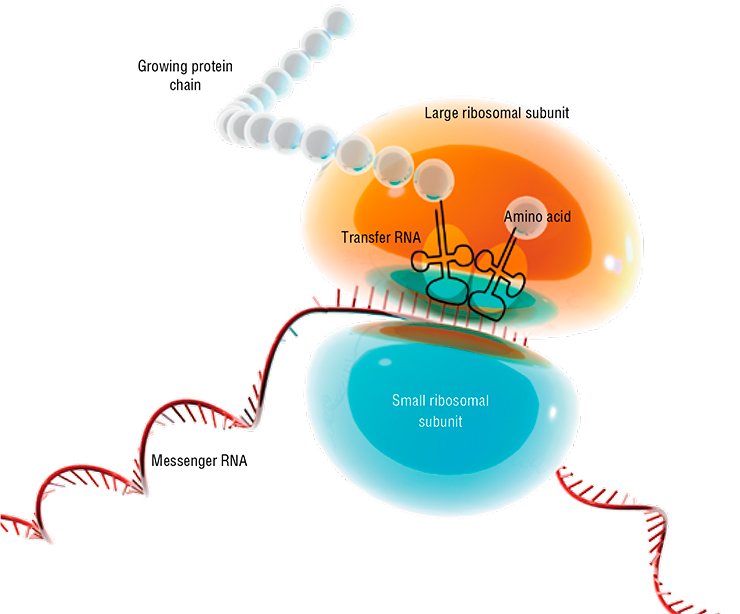
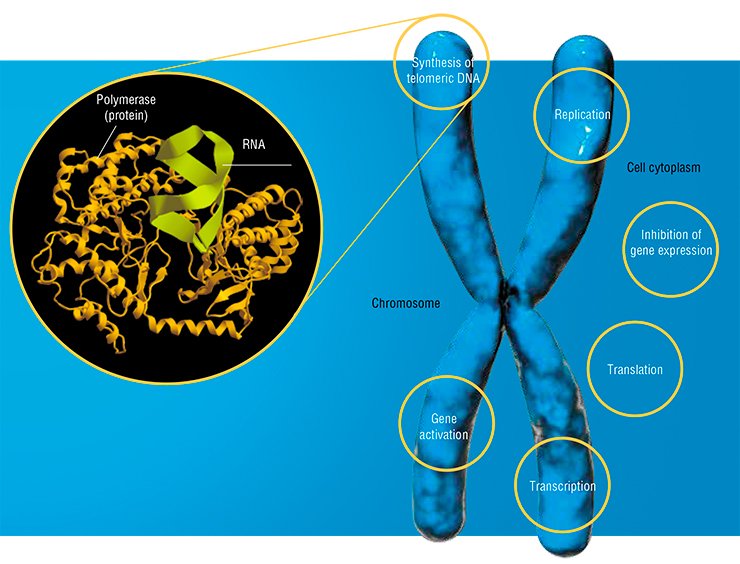
 Moreover, a catalytically active RNA–protein complex, telomerase, was discovered, which appeared to be a main player in the synthesis of the terminal chromosomal regions. As for the RNA contained in the ribosome, it not only plays a merely structural role, but also represents the ribosomal catalytic center! Thus, all the key functions in the process of protein synthesis belong to RNA molecules.
Moreover, a catalytically active RNA–protein complex, telomerase, was discovered, which appeared to be a main player in the synthesis of the terminal chromosomal regions. As for the RNA contained in the ribosome, it not only plays a merely structural role, but also represents the ribosomal catalytic center! Thus, all the key functions in the process of protein synthesis belong to RNA molecules.
It has been shown that, similarly to proteins, RNAs can form compact structures able to interact in a highly specific manner with almost any small and large molecules. RNA catalysts have been found in nature, being in use among both viruses and cells of higher organisms. Quite soon, most diverse catalytic RNAs were produced artificially.
The facts persistently indicated the primacy of RNA as a biological molecule. Indeed, DNA is a stable keeper of information, but it cannot work without the help of proteins. Proteins are efficient catalysts but fundamentally unable to act as information carriers. As for RNA, it has appeared such a universal multifunctional molecule as a true “ancestress” should be.
It has become clear that the currently existing biological world has evolved from such simple RNA-containing systems. But where had this “world of RNA” gone, being replaced by more specialized molecules? Do RNAs today represent a sort of “molecular fossils,” playing although an important but limited role? Fairly recently, the answer to this question seemed completely certain.
Thirty years after
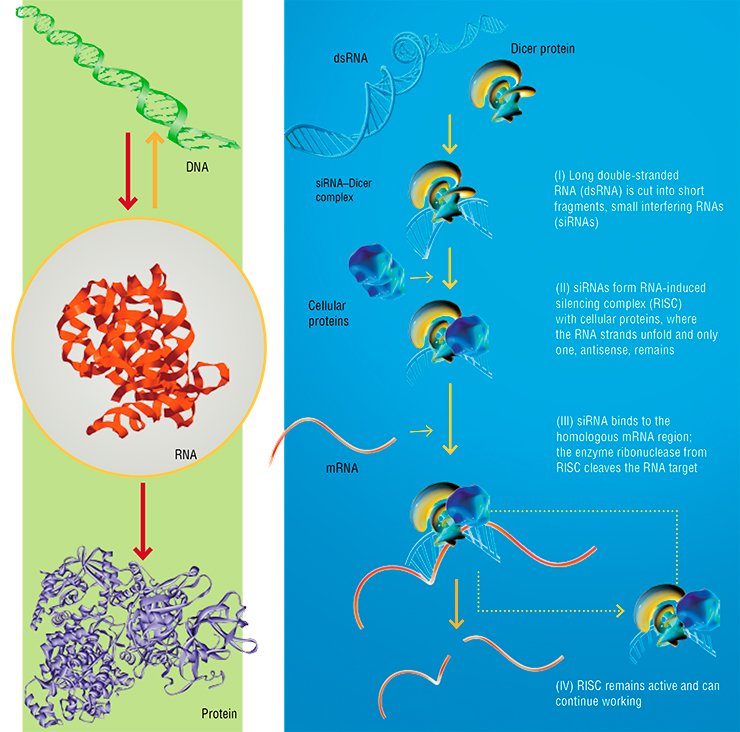
In 1997, a conference in Germany was held on the research into RNA. It was to summarize the results of a large-scale RNA research project: anything associated with these biopolymers at that moment seemed clear and sound. A real “bomb explosion” in this atmosphere was the presentation about the discovery of RNA interference, the previously unknown mechanism involved in the control of gene function with the help of specialized small RNAs.
As a matter of fact, it is quite surprising that the discovery of this rather evident mechanism required so much time, since it had been long known that attachment of a small complementary RNA or DNA fragment sufficient in its size to provide selective binding could halt the work of a targeted genetic program, i.e., could “disable” a particular nucleic acid.
This particular method for selective regulation of the functions of nucleic acids was first proposed as early as the 1960s by Novosibirsk scientists N. I. Grineva, et al., who worked in the team headed by D. G. Knorre, future Academician of the Russian Academy of Sciences. They reasonably decided that the best method to influence a gene in a targeted manner was to utilize the so-called antisense oligonucleotides, that is, short complementary DNA or RNA fragments (Belikova et al., 1967).
 Finally, decades after, it was proved that this mechanism of a targeted action on genetic material actually worked in living systems being involved in the regulation of gene expression. This is a rare instance in the history of science when a phenomenon was first theoretically described, then implemented in experiment, and only decades later discovered in nature.
Finally, decades after, it was proved that this mechanism of a targeted action on genetic material actually worked in living systems being involved in the regulation of gene expression. This is a rare instance in the history of science when a phenomenon was first theoretically described, then implemented in experiment, and only decades later discovered in nature.
Running ahead, we would like to note that Siberian researchers opened with their works a new field in applied molecular biology, currently developing worldwide. The first pharmaceuticals involving interfering microRNAs have been already designed, and creation of a widest range of drugs is expected in future, including the preparations for inactivation of microRNAs themselves (such inhibitors have even got the special name, anti-mir, mir being the Russian word for world) as well as of other noncoding RNAs.
At the same conference of 1997, discovery of numerous manifold noncoding RNAs in cells of the brain and other organs with their concentrations varying depending on the state of the body was reported. It became clear that the RNA research should be further expanded rather than curtailed.
The treasures of the “junk” DNA
Quite soon, RNA interference was found to be most widely spread in nature, and the applied research with interfering RNA aiming to design therapeutics and genetically modified organisms was started.
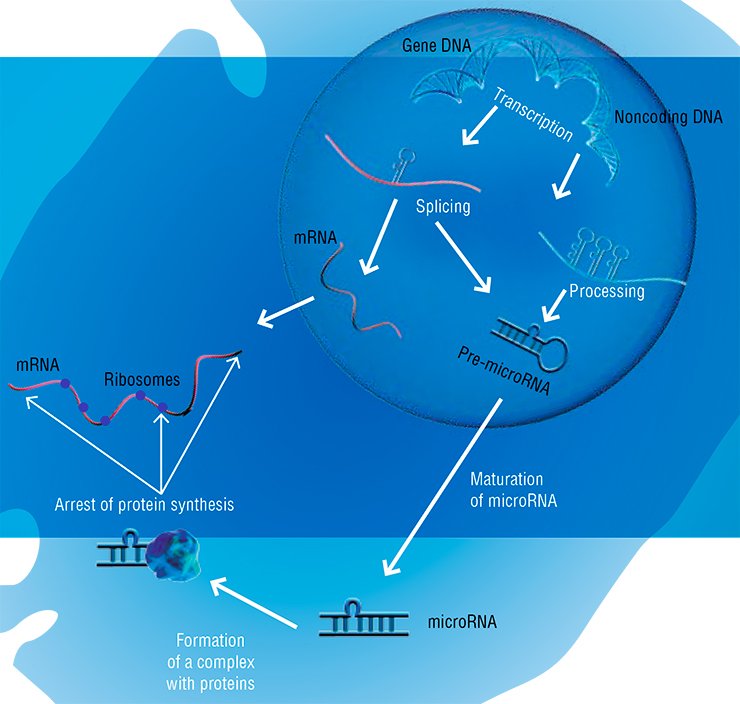
Moreover, it has been shown that organisms synthesize a huge number, thousands of species, of noncoding RNAs, which are encoded in the fabled “junk DNA”. Among the discovered RNA species are the microRNA regulators for gene activities, microRNAs fulfilling various other regulatory functions, as well as many other RNA types with still vague roles. A surprising fact is that many RNAs are localized not only to the cells forming various tissues, but also to the blood, thus being able to “travel” across the body.
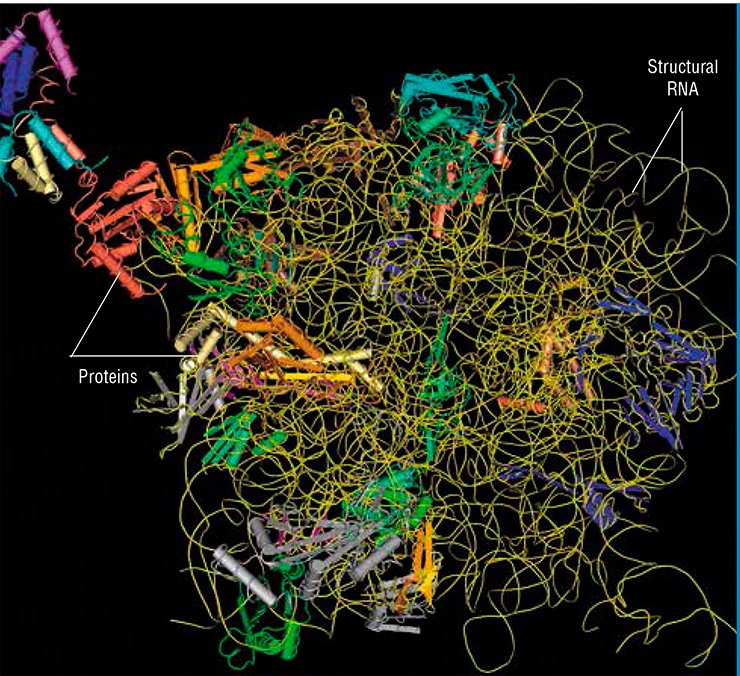 Long noncoding RNAs are still poorly studied, and the mechanisms underlying their function have been clarified only for some cases. Researchers lean to infer that such RNAs play the role of organizers in various complex functional structures comprising different macromolecules. The RNA molecule is an ideal candidate for such a role, since it contains different modules capable of binding both individual proteins and DNA regions. Such RNA modules may be arranged in a definite pattern relative to one another, thereby providing for assembly of supramolecular ensembles of any degree of complexity. The most illustrative example here is a ribosome.
Long noncoding RNAs are still poorly studied, and the mechanisms underlying their function have been clarified only for some cases. Researchers lean to infer that such RNAs play the role of organizers in various complex functional structures comprising different macromolecules. The RNA molecule is an ideal candidate for such a role, since it contains different modules capable of binding both individual proteins and DNA regions. Such RNA modules may be arranged in a definite pattern relative to one another, thereby providing for assembly of supramolecular ensembles of any degree of complexity. The most illustrative example here is a ribosome.
Thus, RNA is the main contributor to the major cell structures that make the cell “live.” In addition, long noncoding RNAs control functioning of cell’s genetic machinery. They switch on and off the work of large chromosomal regions by defining the sites for specific modification of chromosomal proteins.
 The concentrations of diverse microRNAs in organs and tissues are also tightly associated with the state of the organism. Being involved in activity regulation of a tremendous number of genes, these RNAs influence the most important physiological and metabolic processes at the levels of both single cell and overall body.
The concentrations of diverse microRNAs in organs and tissues are also tightly associated with the state of the organism. Being involved in activity regulation of a tremendous number of genes, these RNAs influence the most important physiological and metabolic processes at the levels of both single cell and overall body.
For example, it is possible to use a certain set of microRNAs to change the degree of cell differentiation to obtain undifferentiated stem cells from specialized ones or, on the contrary, to control differentiation of stem cells in a desired direction. As has been proved, microRNAs are involved in the differentiation of the adipose tissue cells, lipid metabolism, secretion of insulin and other hormones, and so on.
The latest data suggest that over 60% of all genes fall within the “jurisdiction” of RNA. Actually, microRNAs are the “managers” governing the development of whole organs and the overall organism by triggering some processes and halting others according to the corresponding biological clocks.
Within the specialized membranous structures involved in transportation, such as exosomes, microRNAs can be transferred not only between tissues and organs of the same organism, but also between individuals. For example, mother’s milk contain exosomes carrying the microRNAs that play an important role in the establishment of the child’s immune system; note that the level of their secretion is especially high during the first six months of lactation (Kosaka et al., 2010).
 It is no surprise that the current interest to such transport structures and the composition of microRNAs or other noncoding RNAs they carry is extremely high, and the recently launched research in this field has been taken up by thousands of scientists from many countries. Moreover, the International Society for Extracellular Vesicles was founded and held its first meeting in the April of 2012, where they announced the establishment of a specialized journal.
It is no surprise that the current interest to such transport structures and the composition of microRNAs or other noncoding RNAs they carry is extremely high, and the recently launched research in this field has been taken up by thousands of scientists from many countries. Moreover, the International Society for Extracellular Vesicles was founded and held its first meeting in the April of 2012, where they announced the establishment of a specialized journal.
Points of application
Thus, noncoding RNAs are the major regulators that control the functions of genes and whole genetic ensembles, and the number of newly known noncoding RNAs is constantly growing. Their concentrations and balance are influenced by the pathological processes taking place in the body (for example, tumorigenesis). This evidently demonstrates that the application of quantitative assays for noncoding RNA is most promising for medical diagnosing, while inhibition of the RNA activity associated with the development of a disease will most likely give birth to a new therapeutic approach.
It is currently known that certain microRNA species can be used to stop the development of prostate or breast cancer (Tavazoie et al., 2008). MicroRNAs are also considered as potential drugs for cardiovascular therapy (Latronico and Condorelli, 2009). The exosomes carrying the microRNAs that circulate in the blood in melanoma and various lung cancer cases are in the process of clinical studies (Rabinowits et al., 2009).
The Siberian Branch of the Russian Academy of Sciences is involved in such research. An original high-throughput RNA sequencing technology, elaborated at the Institute of Chemical Biology and Fundamental Medicine (Siberian Branch, Russian Academy of Sciences, Novosibirsk) has made it possible to rapidly and precisely record the microRNA profiles in various clinical specimens. The data on the microRNA profiles in healthy humans have been already published (Semenov et al., 2012), and recording of the RNA profiles for various pathological states is in progress.
In addition, researchers of the institute have obtained promising experimental data for antitumor and antimetastatic therapy with the enzyme RNase A, which influences the tumor microRNA concentration and profile in the cells and blood serum of sick animals (Mironova et al., 2012).
The Institute of Molecular and Cell Biology (Novosibirsk), recently organized with the Siberian Branch of the Russian Academy of Sciences, is also involved in the studies of microRNA aimed at designing diagnostic methods. In particular, they have determined the expression profiles for several microRNAs in various thyroid tumors.
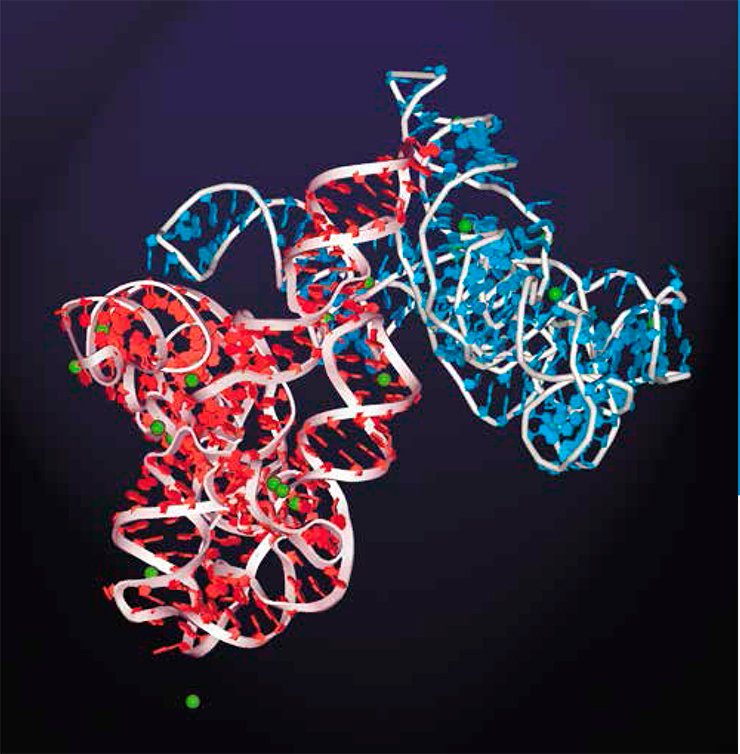 A comprehensive study of the RNA properties prompted researchers to design new technologies promising a power of good to humankind in the nearest future. This is the widely known SELEX technology, allowing for the production of RNA molecules with specified properties, aptamers, which are able to selectively and stably bind the target molecule. In the aptamer technology, the so-called RNA libraries of random sequences containing a great variety of manifold RNA molecules are first created by chemicophysical methods and then this large set is searched for the aptamers using molecular selection according to the ability to specifically interact with the target molecule to produce the found aptamers in the required amounts, although the desired aptamer could be present in the library as a single molecule.
A comprehensive study of the RNA properties prompted researchers to design new technologies promising a power of good to humankind in the nearest future. This is the widely known SELEX technology, allowing for the production of RNA molecules with specified properties, aptamers, which are able to selectively and stably bind the target molecule. In the aptamer technology, the so-called RNA libraries of random sequences containing a great variety of manifold RNA molecules are first created by chemicophysical methods and then this large set is searched for the aptamers using molecular selection according to the ability to specifically interact with the target molecule to produce the found aptamers in the required amounts, although the desired aptamer could be present in the library as a single molecule.
Currently, not only the RNAs with a typical “natural” structure are used as aptamers and therapeutics: various chemical modifications have made it possible to design the molecules resistant to the enzymes destroying RNAs, which are abundant in biological media. Such “artificial” RNAs are most promising in diagnosing; for example, it is possible to produce the RNA aptamers to certain molecules marking a disease and design the biosensors with a unique sensitivity using these aptamers.
The combination of RNA-based bioanalytical approaches to diagnosing diseases and the therapeutic tools involving microRNAs and noncoding RNAs forms the background for a close breakthrough in medicine.
Each year brings the discoveries revealing ever more new facets the miraculous world of RNA. High-throughput technologies for sequencing nucleic acids have allowed a tremendous set of various RNAs to be discovered in cells and blood, including the transcripts of the DNA regions earlier regarded as “silent.”
 Thus, the natural mechanism of paramount importance was discovered at the end of the 20th century; this mechanism controls the operation of cell genome and was forecasted by Siberian scientists long before this discovery as the idea of “antisense nucleotides.”
Thus, the natural mechanism of paramount importance was discovered at the end of the 20th century; this mechanism controls the operation of cell genome and was forecasted by Siberian scientists long before this discovery as the idea of “antisense nucleotides.”
The insight into the mysteries of the world of RNA, which for a long time concealed itself from researchers behind the screen of narrower specialized molecules—proteins and DNA—has brought us closer to understanding of the basic issues in the origin of life. Moreover, this world of RNA, which has taken shape in new promising biomedical technologies, is gradually becoming part in the daily life of humankind.
References
Vlassov, V. V. and Vlassov, A. V. Life Began with RNA // Science First Hand. 2004. No. 2 (3). Pp. 6–19.
Vlassov, A. V. Evolution in a Tube // Science First Hand. 2006. No. 1 (7). Pp. 50–59.
Vlassov, V. V. The Remedy for Genes // Science First Hand. 2007. No. 2 (14). 56–59.
Karpova, G. G., Graifer, D. M., and Malygin, A. A. The Ribosome: A Minifactory for Protein Production // Science First Hand. 2006. No. 6 (12). Pp. 46–53.
Frank-Kamenetsky, M. D. The Uppermost Molecule, Moscow: Nauka, 1983 [in Russian].
Chernolovskaya, E. L. RNA Interference: Fighting Fire with Fire // Science First Hand. 2008. No. 1 (19). Pp. 38–43.
Knorre, D. G., Vlassov, V. V., Zarytova, V. F., et al. Design and Targeted Reactions of Oligonucleotide Derivatives, Boca Raton: CRC Press, 1994.
Vlassov, V. V., Pyshnyi, D. V., and Vorobjev, P. E. Nucleic Acids: Structures, Functions, and Applications. In: Handbook of Nucleic Acids Purification, Ed. D. Liu, Boca Raton: CRC Press, 2009.