In search of the lost genomes
One day, we may then be able to understand what set the replacement crowd apart from their archaic contemporaries, and why, of all the primates, modem humans spread to all comers of the world and reshaped, both intentionally and unintentionally, the environment on a global scale. I am convinced that parts of the answers to this question, perhaps the greatest one in human history, lies hidden in the ancient genomes we have sequenced
The human genome is contained in chromosomes that are present in almost every cell in our bodies. It is composed of approximately 3,2 billion nucleotides. When cells replicate for form germ cell that will contribute to the next generation mutations occur. As a result of these mutations 50 to 200 new substitutions exist in every new individual that is born. These substitutions accumulate in the genome over time to the extent that roughly one nucleotide in a thousand differs between two human genomes today, whereas roughly one nucleotide in a hundred differ between a human and a chimpanzee genome. In addition, duplicated DNA sequences differ both between individuals and between species
Each particular nucleotide site in the genome has its own history that could in principle be traced back through past generations. Such a history can be depicted in the form of a tree showing common ancestors shared with the same site seen in other individuals today. However, in reality, it is impossible to trace the history of a single nucleotide site. Therefore, one generally traces the average history of a segment of the genome, or the entire genome, and depicts that in the form of a tree that thus represents an average picture of how most sites in the DNA segments or genomes whose nucleotide sequences have been determined are related.
Until recently, it was only possible to determine DNA sequences and whole genome sequences from present-day individuals from which DNA can be isolated in good condition from fresh tissues such as blood. To evolutionary scientists this is somewhat frustrating because it represents an indirect way to study the past: one studies DNA sequences that exist today, uses the best models we have for how mutations accumulate and estimates what common ancestors in the past may have looked like. This is frustrating because what we have are estimates subjects to many uncertainties for example as a result of the mutational models used. Since over 30 years, our laboratory works on methods to overcome this “time trap” by going back in time and retrieving DNA sequences from archaeological and paleontological remains. This is possible only on rare occasions when well-preserved tissues can be found. Direct ancestors of present-day organisms are also almost never available. However, this approach nevertheless opens up new possibilities in that it allows DNA sequences from past populations and extinct species to be determined.
 Of particular interest to us is the closest extinct relative of all present-day humans: the Neandertals. This robust form of hominins emerged in Europe and western Asia approximately 300,000 to 400,000 years ago and disappeared between 30,000 and 40,000 ago. The debate concerning the relationships between Neandertals and modern humans and about what happened when they met lasted for decades. One idea was that modern humans replaced Neandertals without interbreeding, in which case the Neandertal contribution to present-day human genetic variation would be zero. Another idea was that Neandertal were the direct ancestors of Europeans. In this case, the Neandertal genetic contribution to present-day people in Europe would approach 100 %. Obviously, all levels of contribution between 0 % and 100 % are also possible, and different levels of Neandertal contribution to present-day Europeans have been argued for on the basis of archaeological and paleontological data.
Of particular interest to us is the closest extinct relative of all present-day humans: the Neandertals. This robust form of hominins emerged in Europe and western Asia approximately 300,000 to 400,000 years ago and disappeared between 30,000 and 40,000 ago. The debate concerning the relationships between Neandertals and modern humans and about what happened when they met lasted for decades. One idea was that modern humans replaced Neandertals without interbreeding, in which case the Neandertal contribution to present-day human genetic variation would be zero. Another idea was that Neandertal were the direct ancestors of Europeans. In this case, the Neandertal genetic contribution to present-day people in Europe would approach 100 %. Obviously, all levels of contribution between 0 % and 100 % are also possible, and different levels of Neandertal contribution to present-day Europeans have been argued for on the basis of archaeological and paleontological data.
We got a first chance to directly test these hypotheses in the mid‑90s when we were allowed to analyze the Neandertal bones that were discovered in the Neandertal Valley in Germany in 1856 and gave its name to this hominin group. At the time, we were able to draw on over ten years of experience with the development of techniques to extract and amplify small amounts of DNA from ancient remains of cave bears, mammoths and other late Pleistocene mammals (Pääbo, 2014). We focused on the mitochondrial DNA (mtDNA), because every cell contains hundreds or even thousands of mtDNA copies, making it easier to retrieve mtDNA than any particular part of the nuclear genome. We reconstructed the most variable part of the mtDNA and estimated phylogenetic trees to reconstruct the history of the mtDNAs of Neandertals and present-day people. In contrast to the nuclear genome, the mtDNA is inherited as one single unit from mothers to offspring without recombination so a phylogenetic tree for mtDNA reflects not the average history but the exact maternal lineages that relate the mtDNA analyzed. On the one hand these trees showed what was already known, that the mtDNAs of all people inhabiting the Earth today trace their ancestry back to a common ancestor about 100,000—200,000 years ago. But they also showed that the mtDNA lineage of the Neandertal type specimen went much further back in time and shared a common ancestor with present-day mtDNAs in the order of half a million years ago (Krings et al., 1997). Subsequently, we and others have determined several other Neandertal mtDNA sequences. They all fall together outside the variation of the mtDNAs of present-day people. Thus, in 1997, it was clear that for the mtDNA, the complete replacement model held: no person today carries an mtDNA derived from a Neandertal.
However, the mtDNA represents only a tiny part of our total genome. The full picture of our genetic history can only be obtained by studying the nuclear genome. In the early years of this millennium it became feasible to consider sequencing genomes from ancient organisms thanks to new techniques that made it possible to sequence millions of DNA molecules rapidly and inexpensively. We were lucky to receive funding from the Max Planck Society for a five-year effort to improve the technique of the extraction of DNA from ancient bones and making DNA libraries that could be used for high-throughput DNA sequencing. We also analyzed a large number of bones from many sites in Europe to find those bones that contained the largest relative proportion of Neandertal DNA. We settled on a site in Croatia, from which we used tree bones from different Neandertal individual and sequenced more than one billion short DNA fragments extracted from the bones. We developed computer algorithms to match these short DNA sequences to the human genome while accounting for errors induced by chemical process that have affected them over tens of thousands of years. Only a few percent of all sequences derived from the Neandertal individuals. Nevertheless, in 2010 we were able to present about 3 billion nucleotides of Neandertal DNA that had been mapped to the human genome. Together these DNA fragments covered about 55 % of the parts of the Neandertal genome to which short fragments can be mapped (Green et al., 2010). This was enough to ask if any genetic interaction had occurred when modern humans encountered Neandertals.
If Neandertals made no genetic contribution to modern humans, the Neandertal genome would be equally far from Africans, Europeans and any other present-day populations. In contrast, if present-day Europeans carried the DNA that they had inherited from Neandertals, European genomes would carry fewer differences to Neandertals than African genomes, since Neandertals were never in Africa so would not be expected to have contributed to genomes there. To test this, we sequenced the genomes of five present-day people and identified positions where two of these differed from each other. We then asked how often at these positions the Neandertal genome carried the variant seen in one present-day person and how often it carried the variant seen in the other present-day person. This approach of counting matches to pairs of present-day genomes was necessary since the quality of the Neandertal genome was so low that we could not trust sequence variants that were seen only in the Neandertal genome and not also in one of the present-day genomes. When we compared two African genomes in this way, the Neandertal genome matched variants in the two genomes equally often. This is to be expected since there was no reason to expect that Neandertals would have contributed DNA to the ancestors of any of the Africans. Intriguingly, when we compared a European and an African to the Neandertal genome, we detected statistically significantly more matching to the European genome, suggesting that Neandertals had contributed DNA to the ancestor of the Europeans. Even more surprising was that when we compared a person from China to an African, and a person from Papua New Guinea to an African, we always found that the non-African matched the Neandertal genome more often than the African genome. This was surprising to us since Neandertals have probably never been in China and surely never in New Guinea. How could this be?

The explanation that we suggested and that has since been borne out by work in our own and other groups was that Neandertals met modern humans and mixed with them probably in the Middle East. If these modern humans later became the ancestors of everybody that today live outside Africa, these early modern humans can so to speak have carried with them the Neandertal genetic contribution also to geographical areas where Neandertals never existed. As a result, between 1 and 2 % of the genomes of every person whose roots are non-African is of Neandertal origin. That the Neandertal component in the genomes of present-day people has since been dated by studies of the extent to which Neandertal-like DNA segments have been broken down to smaller pieces by recombination that happens in each generation (Sankararaman et al., 2012). It has also been confirmed by subsequent studies of a modern human that is about 40,000 years old who carries much larger segments of Neandertal DNA than present-day people since they lived much closer to the time of mixture (Fu et al., 2014).
Of course, it is unlikely that mixing between Neandertals and modern humans happened only in one population and exclusively in the Middle East, but given the data at hand in 2010 this was the simplest explanation of our findings. Further insights were to a large extent limited by the comparatively low quality of the Neandertal genome. This was to be changed thanks to our collaboration with Anatoly Panteleevich Derevianko.
The excavations at Denisova Cave, led by Academician A. P. Derevianko and Professor M. V. Shunkov of the Institute of Archaeology and Ethnography of the Siberian Branch of the Russian Academy of Sciences have generated many fundamental and novel insights into human evolution. One of their crucial finds is a hominin toe bone discovered in 2010. When we applied new, ultra-sensitive methods that my laboratory have developed to extract DNA and produce DNA libraries to this bone, we were able to sequence almost 50-fold more endogenous DNA from this single small bone than from the three bones from Croatia that had been used to produce the first Neandertal genome a few year earlier. This individual turned out to be a Neandertal and its genome was sequenced to a quality higher than most genomes determined from present-day, living people (Prüfer et al., 2014).

Using such high-quality genomic information, it is possible to observe differences between the two genomes that the individual inherited from her father and from her mother. One can thus gauge the extent of variation in the population where the parents of the individual lived. One can also estimate how closely related the mother and the father of the individual were to each other. In the case of the Neandertal from Denisova Cave this yielded an unexpected result. The paternal and maternal genomes had long segments of DNA that were identical. This means that the parents of this individual were closely related. One can estimate that they must have been related at the level of half siblings. When in the future further Neandertal genomes are sequenced to the same high quality as the one from Denisova Cave, it will be interesting to see if this was an unusual situation among Neandertals or if it reflects the social pattern typical of Neandertals.
The high quality of the Neandertal genome from Denisova Cave can also be used to estimate what parts of the genomes of present-day people were inherited from Neandertals. This confirms that everybody outside Sub-Saharan Africa carries between 1 to 2 % of Neandertal DNA. This proportion is slightly larger in East Asia than in Europe, suggesting that additional admixture between Neandertals and modern humans may have happened during the colonization of Asia (Vernot and Akey, 2015). To get a perspective on this, you may recall that we all have one half of our DNA from each of our parent, about 25 % from each grand-parent, about 12 % from our great grandparents, and so on. From an ancestor six generations back we have on average inherited about 1.5 % of our DNA. Thus, from the point of view, of the total amount of DNA people today have inherited from Neandertals it is as if they had a Neandertal ancestor six generations back. However, due to recombination that occurs when new germ cells are formed in each generation, the Neandertal DNA is distributed in much smaller fragments than the DNA you have inherited from your ancestors six generations back. You may also ask how much of the total Neandertal genome exists distributed among people living today. This estimate is still very approximate but it would seem that at least about 40 % of the Neandertal genome can be found in people today.

Amazingly, the high-quality Neandertal genome is not the only great gift that Denisova Cave has given the world. In 2008 a tiny piece of the phalanx of a fifth finger of a child was discovered in the East gallery of the cave. We were privileged to work on this find and were happy to be able to generate first a low quality genome (Reich et al., 2010) and then, as our techniques improved, a high-quality genome from it. In this genome, each position in the part of the genome amenable to mapping short pieces of DNA was covered over 30 times (Meyer et al., 2012). When we compared this genome to other genomes, we were surprised to find that it was neither a modern human nor a Neandertal. It shared a common ancestor with Neandertals but this ancestral population lived about four times further back in time than the oldest ancestral population shared among present-day human populations. After discussions with Academician A. P. Derevianko and his team in Novosibirsk, it was decided to name this new hominin group “Denisovans.” It is the first hominin group described on the basis of a genome sequence rather than a morphological description. Although remains of Denisovans have yet to be found outside the Denisova Cave, we can learn about their history and the history of other hominins by studies of their genome.
Interestingly, in the order of 5 % of the genomes of people that today live in the Pacific, for example, Aboriginal Australians and Papuans, come from Denisovans (Reich et al., 2011), suggesting the ancestors of these populations met Denisovans and sired offspring with them. In addition, about 0.2 % of the genomes of people in Mainland Asia come from Denisovans (Skoglund and Jakobsson, 2011; Prüfer et al., 2014). By comparing the two high-quality genomes of a Neandertal and a Denisovan that have been determined from Denisova Cave, we can also discern gene flow events that have occurred between these two groups and other gene flow events that have affected these two groups differently. A minimum of two additional instances of gene flow can be detected by these comparisons: one from eastern Neandertals into Denisovans, and one from an unknown hominin that diverged a million or more years ago from the human lineage into Denisovans (Prüfer et al., 2014). In addition, recent work shows that early modern humans in Europe mixed with Neandertals when they first arrived there (Fu et al., in press).

The emerging picture is thus a complicated one, where many different hominin groups exchanged genes with each other on what must have been many occasions. Often this exchange was of limited magnitude but it shows that the gene pools of most or even all hominin groups in the Late Pleistocene were open systems that allowed genetic variants to spread from one group to another. One interesting question then becomes of this may have been of functional importance. As yet, we do not know much about this, but I want to bring up a few examples of what has emerged from studies by several groups in the last two years.
One way to ask what functional role Neandertal genetic variants may play in present-day genomes is to ask what the genes are that carry Neandertal variants that have risen to high frequency. The fact that these variants have become frequent today may suggest that they were positively selected in the past. The group of genes that is statistically overrepresented in such genomic segments are keratins, i. e. structural protein in present in skin and hair (Vernot and Akey, 2014; Sankararaman et al., 2014). Thus, it is likely that in the future we will find that some aspect of the morphology or function of skin and hair that is present in some people in Europe and Asia derive from Neandertals.

There are also aspects of metabolism that are affected by Neandertal variants. For example, Europeans but not Asians carry more Neandertal variants than statistically expected of genes involved in the catabolism (Khrameeva et al., 2014). It is not yet known what these variants do but it will hopefully be discovered in the next few years. Interestingly, a variant of the gene encoding a protein that transports lipids across cell membranes and is derived from Neandertals has risen to a frequency of up to 35 % in East Asia and Native Americans. This variant is associated with increased risk to develop type 2 diabetes (SIGMA Consortium, 2014). It may seem surprising that a Neandertal gene variant that confers the risk of disease has become frequent in the population. One may speculate that a variant that causes diabetes today in people who enjoy ample nutrition throughout life may have represented an advantage in a situation of food shortage. Thus, this gene variant may represent a Neandertal adaptation to starvation that in the past was advantageous also in modern humans.
Have Denisovans like the Neandertals contributed functionally to present-day people? Recent work suggests that this is the case. The population in Tibet carries genetic adaptations to life where the partial pressure of oxygen is low, as is the case at high altitudes on the Tibetan High Plateau. The major gene variant responsible for this adaptation affects the number of red cells in blood and occurs at a frequency of about 80 % in Tibet but is very rare elsewhere in Asia. Last year it was shown that gene variant is likely to be inherited from Denisovans (Huerta-Sánchez et al., 2014). Thus, it seems that the gene flow from Denisovans has contributed to making life at the high plateau in Tibet possible. Similarly, there are indications that gene variants important for how the immune system deals with infectious diseases may have been acquired both from Denisovans and from Neandertals (Abi-Rached et al., 2011).
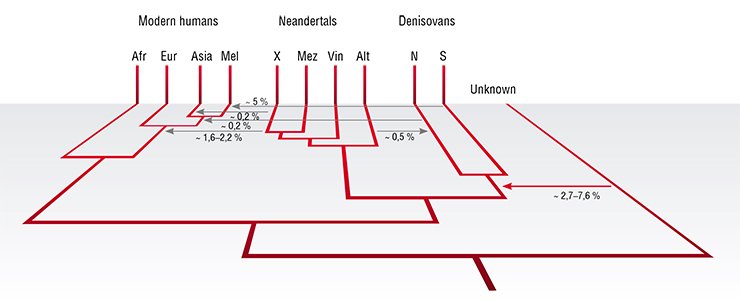
There is thus a picture emerging where Denisovans, Neandertals and possibly other archaic groups who had lived in Eurasia for hundreds of thousands of years and had adapted to local environments met and mixed with modern humans on many occasions. This gave modern humans the opportunity to acquire locally advantageous gene variants from these groups. This is a phenomenon often referred to as “adaptive introgression” in other species (Hedrick, 2013) which may have been of some importance for modern humans as they colonized new environments throughout Eurasia (Racimo et al., in press).
In summary, the fact that a gene flow has been detected not only from Denisovans and Neandertals into modern humans but also between various other hominin groups shows that these were not closed genetic systems. They may best be regarded as a “metapopulation” – a web of populations that included Neandertals, Denisovans, modern humans and other groups, which were linked by but an intermittent or sometimes perhaps even persistent gene flow (Pääbo, in press). In this metapopulation gene variants spread directly, but also potentially indirectly between groups who were in contact with each other over other groups.
These results support the idea expressed by Academician Derevianko already in 2005 when he said “Dear colleagues, please do not offend Neanderthals.
They are among our ancestors!” (Derevianko, 2005). The analyses of the genomes from Denisova Cave have shown that this generous attitude was correct and should be extended to Denisovans and perhaps also other hominin forms.
IN SEARCH OF THE LOST GENOMES* <…> In fact, there is ampleevidence to suggest that genetic traits are a necessary foundation to these human behaviors. In the past, people sometimes did what we now consider to be unethical experiments in which they raised newborn apes together with their own children in their home. Although apes learned how to do many human-like things—they could construct simple two-word sentences, manipulate household appliances, use bicycles, and smoke cigarettes—they did not learn truly complex skills and they did not engage in communication on the scale that humans do. In essence, they did not become cognitively human. So it’s clear that there is a biological substrate necessary for fully acquiring human culture.
<…> In fact, there is ampleevidence to suggest that genetic traits are a necessary foundation to these human behaviors. In the past, people sometimes did what we now consider to be unethical experiments in which they raised newborn apes together with their own children in their home. Although apes learned how to do many human-like things—they could construct simple two-word sentences, manipulate household appliances, use bicycles, and smoke cigarettes—they did not learn truly complex skills and they did not engage in communication on the scale that humans do. In essence, they did not become cognitively human. So it’s clear that there is a biological substrate necessary for fully acquiring human culture.
This is not to say that genes alone are sufficient for acquiring human culture, only that they are a necessary’ substrate. In the imaginary experiment where a human child is raised in the absence of any contact with other human beings, it is very likely that the child would never develop most of the cognitive traits that we associate with humans, including awareness of the interests of others. That unfortunate child would probably also not develop the most sophisticated of cultural traits that emanates from our tendency to share attention with others: language. So I am convinced that social input is necessary for the development of human cognition. However, no matter how early in life and how intensively they are integrated into human society and no matter how much teaching they are subject to, apes do not develop more than rudimentary cultural skills. Social training alone is not enough. A genetic readiness to acquire human culture is necessary. Similarly, I am convinced that a newborn human raised by chimpanzees would fail to become cognitively chimpanzee. There is surely also a genetic substrate necessary to becoming fully chimpanzee that humans lack. But since we are humans, we are more interested in what makes humans human than in what makes chimpanzees chimpanzee. We should not be ashamed of being “humancentric” in our interests. In fact, there is an objective reason to be so parochial. The reason is that humans, and not chimpanzees, have come to dominate much of the planet and the biosphere. We have done so because of the power of our culture and technology; these have allowed us to increase our numbers vastly, to colonize areas of the planet that otherwise would not have been habitable for us, and to have an impact on and even threaten aspects of the biosphere. Understanding what caused this unique development is one of the most fascinating, perhaps even one of the most pressing, problems that scientists face today. One key to the genetic underpinnings of this development may well be found through comparing the genomes of present-day humans with Neanderthals. Indeed, it is this feeling that kept me going during years of struggling with the technical minutiae of retrieving the Neanderthal genome.
<…>
The dirty little secret of genomics is that we still know next to nothing about how a genome translates into the particularities of a living and breathing individual. If I sequenced my own genome and showed it to a geneticist, she would be able to say approximately where on the planet I or my ancestors came from by matching variants in my genome with the geographic patterns of variants across the globe. She would not, however, be able to tell whether I was smart or dumb, tall or short, or almost anything else that matters with respect to how I function as a human being. Indeed, despite the fact that most efforts to understand the genome have sprung from efforts to combat disease, for the vast majority of diseases, such as Alzheimer’s, cancer, diabetes, or heart disease, our current understanding allows us only to assign vague probabilities to the likelihood that an individual will develop them. So in my realistic moments, I realized that we would not be able to directly identify the genetic underpinnings of the differences between Neanderthals and modem humans. There would be no smoking gun to be found.
Still, the Neanderthal genome was a tool that would allow us to begin to ask questions about what set Neanderthals and humans apart—a tool that not only we but all future generations of biologists and anthropologists would be able to use. The first step was obviously to make a catalog of all the genetic changes that happened in the ancestors of people living today after they separated from the ancestors of the Neanderthals. These changes would be many, and most of them would be without great consequences, but the crucial genetic events that we were interested in would be hidden among them.
The crucial task of making the first version of such a catalog of all changes unique to modern humans was taken on by Martin Kircher together with his supervisor Janet Kelso. Ideally, such a catalog should contain all genetic changes that are present today in all or nearly all humans and that occurred after modem humans parted ways with the ancestors of Neanderthals. The catalog should thus list positions in the genome where the Neanderthal looked like the chimpanzee and other apes while all humans, no matter where they lived on the planet, differed from the Neanderthals and the apes. However, in 2009 there were many limitations to how complete and correct such a catalog could be. First of all, we had sequenced only about 60 percent of the Neanderthal genome so the catalog could only be 60 percent complete. Second, even if we saw a difference from the human reference genome at a position where the Neanderthal genome looked like the chimpanzee genome, this did not necessarily mean that all humans today looked like the human reference genome. In fact, most such positions would vary among humans, but our knowledge about genetic variation among humans was too incomplete to differentiate real finds and false positives. Fortunately, there were several large projects under way aimed at describing the extent of genetic variation among humans, including the 1,000 Genomes Project, the goal of which was detecting all variants in the human genome present in 1 percent or more of humans. But that project was just starting. A third apparent limitation was that our genome was a composite of sequences from only three Neanderthals, and for most positions, we had only the sequence of a single Neanderthal individual. However, I didn’t view this as overly problematic. As long as one single Neanderthal had the ape-like, ancestral version at a given position, it didn’t matter if other Neanderthals that we hadn’t sequenced carried the derived, new version that we saw in humans today. The knowledge that the ancestral variant was in at least one Neanderthal told us that it had still been around when Neanderthals and modem humans parted ways, perhaps 400,000 years ago. This made it a potential candidate for defining what might be universally modem human.
<…>
Janet and Martin compared the human reference genome with the chimpanzee, orangutan, and macaque genomes and identified all positions where they differed. They then compared all four genomes to our Neanderthal DNA sequences, being careful to compare only those Neanderthal DNA sequences for which we had complete certainty as to where they came from in the genome. They found that we had Neanderthal sequence coverage for 3,202,190 positions where nucleotide changes had occurred on the human lineage. For the vast majority of these positions, the Neanderthals looked like us, which was not surprising, given that we are much more closely related to Neanderthals than to apes. But for 12.1 percent of these positions, the Neanderthal looked like the apes. They then checked whether the ancestral variants seen in apes and Neanderthals were still present in some humans today; in most cases they found both the ancestral and the new variants in present-day humans. This was not surprising because the mutations responsible happened quite recently. But some of these new variants were, as far as we could tell, present in all humans today. These were the positions that we found particularly interesting.
<…>
Clearly our paper was reaching a broader audience than we had ever imagined. But most people weren’t shocked by the idea that their ancestors had interbred with Neanderthals. In fact, many seemed to find the idea intriguing—some, as had happened before, even volunteering to be examined for Neanderthal heritage. By early September, I started to notice a pattern: it was mostly men who wrote to me. I went back through my e-mails and found that forty-seven people had written to say they thought they were Neanderthals—and of these, forty-six were men! When I told my students, they suggested that perhaps men were more interested in genomic research than women. But that didn’t seem to be the case, as twelve women had written to me not because they thought they were Neanderthals but because they thought their spouses were! Interestingly, not a single man had written to make such a claim about his wife (since then, however, one man has actually done so). I joked that some interesting genetic inheritance patterns were at work here that we needed to investigate. But what we were obviously seeing were the effects of the cultural ideas about what Neanderthals were like. In popular lore, Neanderthals are big, robust, muscular, somewhat crude, and perhaps a little simple. Some of these characteristics might be seen as acceptable or even positive in men, but they were clearly not conventionally seen as attractive in women. This idea was brought home to me when Playboy magazine called to ask for an interview about our work. I accepted, thinking that this would probably be my one and only chance to appeal- in Playboy. The magazine ended up writing a four-page story called “Neanderthal Love: Would You Sleep with This Woman?” The accompanying illustration was of a sturdy, very dirty woman wielding a spear on a snowy mountain ridge. That distinctly unattractive image probably explains why hardly any men volunteer the opinion that they are married to Neanderthals.
<…>
 On December 3, 2009, I was attending a meeting on the rat genome at the Cold Spring Harbor Laboratory. I was there to describe a project on artificial domestication in rats that my group had been working on for the last few years. As I walked from the dining hall to the lecture hall after breakfast, my cell phone rang. It was Johannes Krause calling from Leipzig and he sounded strangely excited. I asked him what the matter was. He asked me if I was sitting down. When I said no, he said I’d better sit down before hearing what he had to tell me. Stalling to worry that something terrible had happened. I sat down.
On December 3, 2009, I was attending a meeting on the rat genome at the Cold Spring Harbor Laboratory. I was there to describe a project on artificial domestication in rats that my group had been working on for the last few years. As I walked from the dining hall to the lecture hall after breakfast, my cell phone rang. It was Johannes Krause calling from Leipzig and he sounded strangely excited. I asked him what the matter was. He asked me if I was sitting down. When I said no, he said I’d better sit down before hearing what he had to tell me. Stalling to worry that something terrible had happened. I sat down.
He asked me if I remembered a small bone that we had gotten from Anatoly Derevianko in Russia. <…>
Some years previously, Anatoly had visited our laboratory and given us a few small bones in plastic bags. They had been excavated in a spot called Okladnikov Cave in the Altai Mountains in southern Siberia, where Russia, Kazakhstan, Mongolia, and China meet. These bones from Okladnikov Cave were too fragmentary to tell what type of human they had come from, but we extracted DNA from them and showed that they contained Neanderthal mtDNA. Together with Anatoly, we then published a paper in Nature in 2007 that extended the range where Neanderthals had lived by at least 2,000 kilometers further east of what had been commonly believed. Prior to our paper, no Neanderthal had been confirmed east of Uzbekistan.
In the spring of 2009 we received another bone fragment from Anatoly. His team had discovered that fragment during the previous year in Denisova Cave, another cave in the Altai region located in a valley that connects the Siberian steppes in the north to China and Mongolia in the south. The bone was minuscule, and I hadn’t attached very much importance to it, thinking only that we would see whether it contained any DNA at some point in the future when there was time. Perhaps it would prove to be Neanderthal, which would enable us to gauge the extent of mtDNA variation among the easternmost Neanderthals.
Johannes had now found the time to extract DNA from the bone; and Qiaomei Fu, a talented young graduate student from China, had made a library and used a method that Adrian Briggs, the British graduate student in our lab. had developed to fish out mtDNA fragments from the library. They found a very large amount of mtDNA—in total 30,443 fragments, which enabled them to assemble the complete mitochondrial genome with a very high degree of accuracy. In fact, each position in the mtDNA was seen an average of 156 times, unusually high for an old bone. That was good news, but it wasn’t why Johannes asked me to sit down. He had compared the mtDNA sequence of the Denisova bone to the six complete Neanderthal mtDNA sequences that we had previously determined as well as to mtDNA sequences from present-day humans from around the world. Whereas the Neanderthals differed from modem humans at an average of 202 nucleotide positions, the Denisova individual differed at an average of 385 positions—almost twice as much! In a tree analysis, the Denisova mtDNA lineage branched off well before the modem human and Neanderthal lineages shared a common ancestor. When Johannes calibrated the rate of substitutions by assuming that humans and chimpanzees split 6 million years ago. then the Neanderthal mtDNA split from the human lineage about half a million years ago—just as we had previously shown—and the mtDNA of the Denisova bone branched off approximately 1 million years ago! I could hardly believe what Johannes was telling me. This was neither a modem human nor a Neanderthal! It was something else, entirely.
My head was spinning. What extinct human group could have split off from the human lineage a million years ago? Homo erectus? But the oldest H. erectus fossils outside Africa were found in Georgia and were about 1.9 million years old. So H. erectus were supposed to have left Africa and thus to have split from the lineage leading to present-day humans almost 2 million years ago. Homo heidelbergensis? But they were thought to be the direct ancestors of Neanderthals and would then presumably have diverged from the modem human lineage at the same time as Neanderthals. Was this bone from something totally unknown? A new form of extinct human? I asked Johannes to tell me everything about this bone.
The bone was indeed tiny, the size of two grains of rice put together. It came from the last phalanx of the little finger, the outermost part of a pinky, from what was probably a young individual. Johannes had used a dentistry drill to remove thirty milligrams of material from the bone, and from this tiny amount of bone powder he had extracted the DNA that Qiaomei had used to make the library. Given how much mtDNA she and Johannes found, the DNA preservation in the bone must be exceptionally good. I would be back in Leipzig in three days and I told him that we would meet then and decide what to do.
After I hung up, I couldn’t bring myself to listen to presentations about how the genomes of different rat strains differed from each other. It was a sunny and snowless winter day in the New York area. I spent the morning walking along the windy beach below Cold Spring Harbor and thought about the young person who had died far away in a Siberian cave many thousands of years ago. All that remained of that life was a tiny speck of bone, but it was enough to tell us that she represented something unknown to us, a group of humans who had left Africa before the ancestors of the Neanderthals but after Homo erectus. Could we find out what this group was?
<…>
As soon as we were back in Leipzig we finished the manuscript, which we entitled “The Complete mtDNA Genome of an Unknown Hominin from Southern Siberia,” and sent it off to Nature. It was a unique paper. For the first time ever, a new form of extinct humans was described from DNA sequence data alone, in the total absence of any skeletal remains. Given that the mtDNA was so different from that of both modem humans and Neanderthals, we felt sure that we had found a new form of extinct human. In fact, we were so taken with this idea that, after some discussion, we decided to describe it as a new species, which we called Homo altaiensis.
However, I felt vaguely uneasy about suggesting a new species and soon had second thoughts. To me, taxonomy, the classification of living organisms into species, genera, orders, and so on, is a sterile academic exercise, particularly when discussing extinct human forms. Whenever my students send me manuscripts in which they use Linnaean Latin names for groups that are commonly known for example, “In order to better understand the pattern of genetic variation in Pan troglodytes, we sequenced…”
I always delete the Latin and sometimes even snidely ask who they are trying to impress by saying “Pan troglodytes” instead of “chimpanzees.” Another reason I dislike taxonomy is that it has a tendency to elicit scientific debates that have no resolution. For example, if researchers refer to Neanderthals as “Homo neanderthalensis,” they indicate that they regard them as a separate species, distinct from “Homo sapiens.” This invariably infuriates multiregionalists, who see continuity from Neanderthals to present-day Europeans. If researchers say. “Homo sapiens neanderthalensis,” they indicate that they see them as a subspecies, on par with “Homo sapiens sapiens.” This invariably infuriates proponents of the strict out-of-Africa hypothesis. These arguments I prefer to avoid, and although we had by now shown (but not yet published) that there had been mixing between Neanderthals and modem humans. I knew that taxonomic wars over Neanderthal classification would continue, since there is no definition of a species perfectly describing the case. Many would say that a species is a group of organisms that can produce fertile offspring with each other and cannot do so with members of other groups. From that perspective we had shown that Neanderthals and modem humans were the same species. However, this concept has its limitations. For example, polar bears and grizzlies can (and occasionally do) produce fertile offspring with each other when they meet in the wild. Yet polar bears and grizzlies look and behave differently, and are adapted to different lifestyles and environments. It would seem rather arbitrary, if not outright ridiculous, to regard them as one and the same species. We didn’t know whether the fact that Neanderthals contributed perhaps 2 to 4 percent of the genes of many present-day humans meant that they were the same or different species. So it was ironic that, having always refrained from using a Latin name for Neanderthals in our papers, I was now on the verge of introducing a new Limiaean species designation myself.
Despite my misgivings about fruitless taxonomic debates, I felt I had some reasons for this digression from my principles. The mtDNA of the Denisova individual was about twice as different from the mtDNAs of modem humans as was the mtDNA of Neanderthals. That probably made them more like H. heidelbergemis, who did get to have their own Latin species name. But there was also vanity involved. Not many people get to name a new hominin species, which made it tempting to do so, even more so because this was the first time it would be done based solely on DNA data. However, the deciding argument came both from some people in our group and from Henry Gee at Nature. He pointed out that if we didn’t take the initiative and give this hominin group a species name, someone else would. And that person might come up with a name we didn’t like. So, after deliberating with Anatoly and the team who had excavated the crucial finger bone, we settled on provisionally naming it Homo altaiensis.
Nature kept its promise to process our paper quickly: eleven days after our submission, we received comments from four anonymous reviewers. They all praised the technical aspects of the paper but they were divided on the issue of naming a new species. Two reviewers voiced concerns that we might actually have sequenced a late Homo erectus. They felt that if H. erectus had had continuous contact with groups in Africa, they may not show an mtDNA divergence as deep as their first exit out of Africa some 2 million years ago. I doubted this. But the fourth reviewer made the point that saved us from ourselves. He or she said that “once a name is in the taxonomic literature, it cannot be withdrawn later. So such provisional naming is not wise, I believe.” When I read this, I realized we had been foolish.
In the meantime, it dawned on us that the very large amounts of mtDNA that Johannes had been able to capture from the Denisova DNA libraries meant we would be able to sequence quite a bit of this individual’s nuclear genome. This would settle its relationships both to Neanderthals and to modem humans in a definitive way as well as its possible status as a new species. We rewrote the manuscript and removed any reference to a new species. Instead, we said that “nuclear DNA sequences are needed to clarify definitively the relationship of the Denisova individual to present- day humans and Neanderthals.” We sent it back to Nature, where it appeared in early April. As events would show, we had reason to be grateful that we had not named it a new species.
<…>
Monty Slatkin used all the DNA sequences we had generated to test various population models. As I expected, he found that the simplest model that explained all the data was admixture between Neanderthals and modem humans, followed by later admixture between Denisovans and Melanesian ancestors. But we still needed to explain the very strange Denisovan mtDNA. There were two possibilities. One was that the mtDNA lineage was introduced into Denisovan ancestors through admixture with another, more archaic hominin group. This was the idea I secretly favored. The other was that it was due to a process known as “incomplete lineage sorting.” This means simply that the population that was the common ancestor of Denisovans and Neanderthals as well as modem humans carried earlier versions of all three mtDNAs. Then, by chance, one mtDNA variant that carried a lot of differences from the other two became the one that survived in Denisovans whereas the other two, which were much more similar to each other, became the ones that survived in Neanderthals and modem humans, respectively. This was particularly likely to have occurred if the ancestral population of Denisovans, Neanderthals, and modem humans was large enough that many mtDNA lineages could have coexisted in it. Monty’s population models showed that the data could be explained either by a small amount of admixture from another unknown human group or by this “incomplete lineage sorting” scenario. Although that meant we couldn’t favor one explanation over another, admixture nonetheless seemed a more plausible explanation to me. After all, we had already detected two cases of mixture between archaic groups and modem humans, so I had become much more open to the possibility that mixing was a common feature during human evolution. Furthermore, if the Denisovans were willing to have sex with modem humans, it seemed plausible that they would have sex with other archaic groups as well. I had come to believe that although the big picture of modem human spread was one where the replacement crowd pushed other groups into extinction, this was not a total replacement. Rather, some DNA seemed to leak over into the groups that lived on, so much so that I started using a term I had picked up from somewhere to describe this process: “leaky replacement.” Perhaps, I thought, the spread of Denisovans had also been a “leaky” affair.
<…>
But even though we had sequenced the Neanderthal genome and opened the door to the genomes of other extinct human groups, many mysteries remained. One big mystery was when the Denisovans had lived. Both the finger bone fragment and the tooth were too small to allow- us to obtain radiocarbon dates. Instead, we had dated seven bone fragments, most with cut marks or other human modifications, found in the same layer in Denisova Cave. Four of the seven turned out to be older than 50,000 years, while three were between 16,000 and 30,000 years old. So it seemed there had been humans in the cave before 50,000 years ago and then again after 30,000 years ago. I tended to think that the older people were the Denisovans and the younger people modem humans, but we couldn’t be sure. Professor Shunkov and Anatoly had found amazingly sophisticated stone tools and a polished stone bracelet in what seemed to be the same layer as the finger bone. Could they have been made by the Denisovans? It was an outlandish idea but the archaeologists felt it was possible.
Another big mystery was how far the Denisovans had ranged. We knew that they were in southern Siberia, but the fact that they had met and conceived children with the ancestors of Melanesians suggested that they had been much more widespread in the past. Perhaps they had roamed all over Southeast Asia, from temperate or even subarctic regions to the tropics. I thought we needed to look for Denisovan DNA in fossils from China. It would also be extremely exciting if Anatoly and his team could find more complete remains of Denisovans in the Altai Mountains. If those bones had features that set Denisovans apart from other hominin groups, these features would perhaps allow us to identify other fossils elsewhere in Asia as Denisovans.
My group and others have since gone on to work on these mysteries. Still other groups have begun to use ancient DNA to study past human epidemics and prehistoric civilizations. But that December I felt a satisfaction rare in my scientific career. What started as a secret hobby when I was a graduate student in my native Sweden over thirty years ago had resulted in a project that seemed like science fiction when we announced it a little over four years earlier. We had now brought this project to a successful conclusion. With my family in our cozy little Swedish hut. I was more relaxed over those Christmas holidays than I had been for a long time.
Acknowledgements. Our work is made possible by the long-standing, fruitful and stimulating collaboration with Academician Derevianko and Prof. Shunkov who lead the work at Densiova Cave. I am also indebted to past and present members of our research team and collaborators who have made the analyses of archaic genomes possible; to the Max Planck Society for continuous support and to the Russian Academy of Sciences for its support of the important work in Siberia and for the honor bestowed upon me today.
References
Abi-Rached L. et al. The Shaping of Modern Human Immune Systems by Multiregional Admixture with Archaic Humans // Science. 2011. Vol. 334, N 6052. P. 89–94.
Green R. E. et al. A Draft Sequence Neanderthal Genome // Science. 2010. Vol. 328. P. 710–722.
Derevianko, A. P. The Middle to Upper Paleolithic Transition in Eurasia: Hypotheses and Facts. Novosibirsk: Izdat. IAET SB RAS, 2005. P. 5–19.
Fu Q. et al. Genome sequence of a 45,000–year-old modern human from western Siberia // Nature. 2014. Vol. 514. P. 445–449.
Hedrick P.W. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation // Molecular Ecol. 2013. Vol. 22. P. 4606–4618.
Huerta-Sánchez E. et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA // Nature. Vol. 512, N 7513. P. 194–197.
Khrameeva E. et al. Neanderthal ancestry drives evolution of lipid catabolism in contemporary Europeans // Nature Communications. 2014. Vol. 5. Doc. 3584.
Krings M. et al. Neandertal DNA sequence and the origin of modern humans // Cell. 1997. Vol. 90. P. 19–30.
Meyer M. et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual // Science. – 2012. – Vol. 338. – P. 222–226.
Pääbo S. Neanderthal Man. Search of Lost Genoms. Basic Books, N.Y., 2014. 288 p.
Pääbo S. The diverse origins of the human gene pool //Nat. Rev. Genet. 2015. Vol. 16. N 6. P. 313—314.
Prüfer K. et al. The complete genome sequence of a Neanderthal from the Altai, Mountains // Nature. 2014. Vol. 505. N 7481. P. 43–49.
Racimo F. et al. Evidence for archaic adaptive introgression in humans //Nat. Rev. Genet. 2015. Vol. 16. N 6. P. 359—371.
Reich D. et al. Genetic history of an archaic hominin group from Denisova cave in Siberia // Nature. 2010. Vol. 468. P. 1053–1060.
Reich D. et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania // The American J. of Human Genetics. 2011. Vol. 89. P. 516–528.
Sankararaman S. et al. The date of interbreeding between neandertals and modern humans // PLoS Genet. 2012. Vol. 8. doi: 10.1371/journal.pgen.1002947
Sankararaman S. et al. The genomic landscape of Neanderthal ancestry in present-day humans // Nature. 2014. Vol. 507. P. 354–357.
Skoglund P., Jakobsson M. Archaic human ancestry in East Asia // Proceedings of the National Academy of Sciences of USA. 2011. Vol. 108. P. 18301–18306.
Vernot B., Akey J. M. Resurrecting surviving Neandertal lineages from modern human genomes // Science. 2014. Vol. 343. P. 1017–1021.
Vernot B., Akey J. M. Complex history of admixture between modern humans and neandertals // Am. J. Hum. Genet. 2015. Vol. 56. P. 448–453.
Williams et al. The SIGMA Type 2 Diabetes Consortium // Nature. 2014. Vol. 506. P. 97–101.













