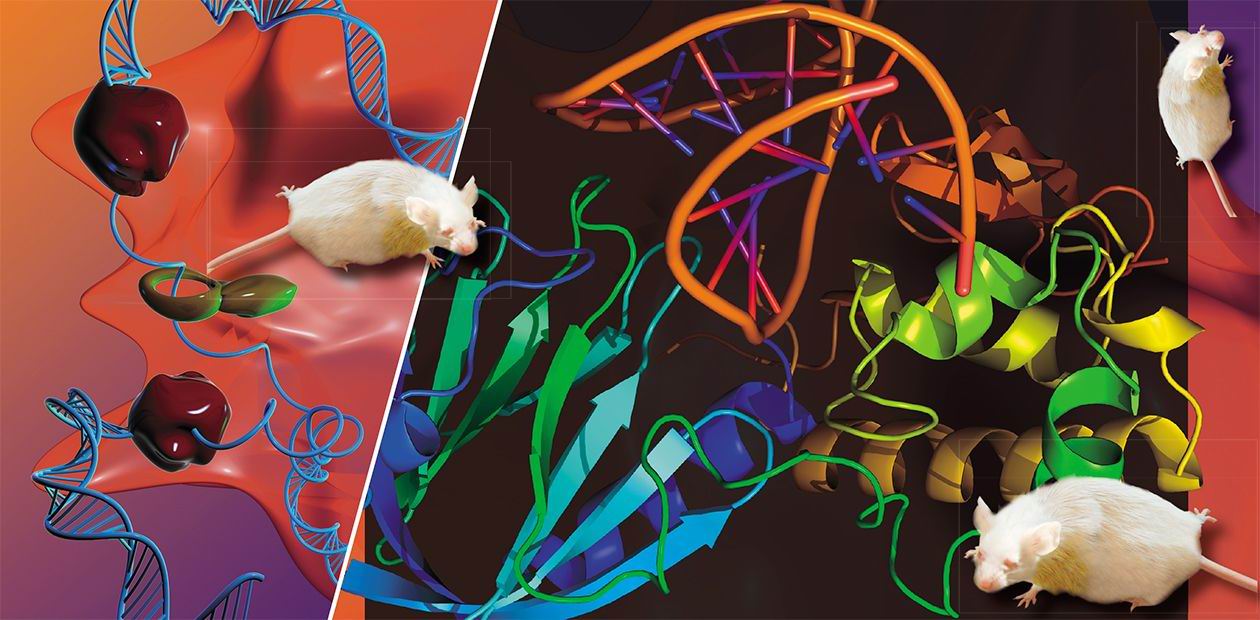
The Enigma of "Rusty" DNA
DNA, the store of genetic information, is vulnerable to a host of environmental factors, including sunlight, ionizing radiation and even the products of cell metabolism. It is hard to imagine what life would be if it were not for dedicated proteins that patrol each cell’s DNA, guarding it from mutations...
It’s hard to tell now when exactly this misfortune happened to the British royal dynasty. Maybe it came in the end of 1737 when Frederick, the Prince of Wales, and his wife Augusta of Saxe-Gotha were conceiving the heir to the throne, or maybe much earlier. The cause is also lost in time: was it a high-energy cosmic ray that hit a piece of royal DNA, or an enzyme molecule that stumbled for a wink and made an error in a germ line cell?
What we do know is the consequences, which resonated all over the world. The baby who was to become King of England George III, though born prematurely, was normal; however, his problems surfaced later. His Majesty suffered from excruciating abdominal pains, tachycardia and neurosis, sometimes going insane for months. While having one of his fits of madness, he signed the act to impose a tea import tax for the American colonies, the spark that ignited the Boston tea party and, later, the American Revolution. Modern physicians have definitely diagnosed King George’s illness as porphyria variegata, a hereditary disease caused by a defect in one of the genes in the hemoglobin biosynthesis pathway. The mutant enzyme works poorly, hemoglobin is made slowly and the accumulating biosynthesis intermediates poison the organism.
So this is how just a single mutation changed the world map and the balance of power. Another widely known example is provided by hemophilia, the inability of blood to clot, which crippled Russian Prince Alexei, thus contributing to the ultimate fall of the Romanov dynasty. Mutations running amok are a terrifying thought indeed. Political trouble aside, life on Earth as we know it would surely be impossible. Fortunately, mutations are rare events.
Plug and socket
Erwin Schroedinger, the famous physicist who wrote the book “What is life?”, boldly stated that hereditary material is so unbelievably chemically stable that it is nearly impossible to change anything in it. Now we know Schroedinger was wrong. If you imagine the genetic material of a single human cell as the Trans-Siberian Railroad, Moscow to Vladivostok, then the number of damages our DNA suffers daily amounts to one breakdown for every hundred yards of the railroad track.
No blame to be put on Schroedinger: his book was written in 1944, preceding the discovery of DNA structure by almost ten years. In fact, DNA is not mentioned in the book at all. Besides, one can hardly expect a physicist to be acquainted with mundane works of many biologists who, at that time, were building the foundation of today’s concepts concerning the mechanisms of mutagenesis. So, he expressed the general knowledge of that time and, as it often happens, made a blunder.
It follows that the cell can fix most of the damage occurring in its hereditary material. This process, discovered in the late 1940s, has been actively studied since then; it is termed DNA repair.
To understand this mechanism, let’s first recall something from high school. The information in DNA is written in four “letters”: A, G, T and C. Of course, these letters are in reality chemical moieties, the nucleobases called adenine, guanine, thymine and cytosine, respectively. They can form bonds with each other, being exactly complementary, just as a plug and a socket. That’s why, in DNA, A always stands opposite T, and G, opposite C. An attempt to put, say, A opposite C, is not quite unlike an attempt to force a European round-pin plug into an American flat-hole socket.
The cell also possesses dedicated enzymes, DNA polymerases, which can “read” one strand of DNA and assemble a complementary strand on this template. Obviously, the sequence of the template strand unambiguously determines the sequence of the complementary strand, providing the basis for the exact copying of hereditary information between generations.
Now, suppose one letter in the strand gets awry, just like if we tape up one hole in the socket. The DNA polymerase will then go in doubt over what to insert opposite the wrong letter. If it puts the correct, complementary letter, everything will stay in order; but if the incoming letter is wrong, a change in DNA will occur, resulting in a mutation. This is not the only mechanism of mutagenesis, but one the most relevant to the following discourse.
Special proteins patrol DNA strands like railroad trackwalkers, searching for lesions.In their famous 1953 paper in Nature, James Watson and Francis Crick, describing the structure of DNA, discussed the role of two complementary DNA strands in the faithful transmission of genetic information but missed an important point, although later Watson claimed it was too trivial to deserve a mention. The point in question is this: If a nucleobase is damaged, there is always a possibility to fix it using a complementary base in the undamaged strand as a template.
In most cases, the repair exploits this very possibility. A number of proteins always patrol DNA like railroad trackwalkers, who keep an eye on the rails. If they find a non-canonical nucleobase, they excise it; then the gap is filled, with the undamaged strand used as a template.
The reason for rusting
How, then, can DNA “letters” be broken? Like with any complex construction, there are a lot of ways to do it. Many of us have heard about the harm done by ultraviolet light: indeed, cross-links that form between adjacent “letters” of UV-irradiated DNA interfere with the proper work of DNA polymerases. However, solar UV irradiation does not illuminate us humans any deeper than the uppermost skin layer. Ionizing radiation can also damage DNA. Nevertheless, the main culprits are associated with the processes of utmost importance for our lives. Among the worst offenders is oxidative DNA damage.
Ionizing radiation is among the agents damaging our DNA. Its main source is radon gas, seeping out of the Earth’s interior everywhere. Other natural radioactive isotopes (mostly the potassium isotope 40K) can also accumulate in the body but contribute much less to the overall irradiation. Cosmic rays are not too dangerous if you are not a frequent flyer; however, they are a significant factor for airplane pilots, more than twice exceeding the impact of radon.In addition to this unavoidable natural background, we also encounter man-made sources of ionizing radiation, the most significant of which is medical X-ray imaging. Minuscule quantities of radioactive isotopes can be found in certain consumer goods, such as smoke detectors. Building materials leak radon, too: its concentration is higher in houses built of reinforced concrete as compared with brick houses (unless they’re made of red Flemish brick), while emission from wood is negligible.
On the early Earth with its mostly oxygen-free atmosphere, UV light was DNA’s enemy number one: the ozone shield had not appeared yet, and the single-celled organisms, unlike us, could be “lighted through”. At that time, aerobic respiration was a thing unheard of: metabolism was based on fermentation or other chemical processes rather than on the reduction of molecular oxygen. Gradually, thanks to photosynthetic organisms, oxygen concentration in the atmosphere rose until some 2 billion years ago it reached the so-called Pasteur point (around 1 %). This was still only one-twentieth of today’s level but already enough for efficient oxygen-based metabolism. The problem is that oxygen is a very strong oxidizing agent; therefore, special efforts are necessary to steer the reactions with oxygen in the required direction while avoiding the stray ones.
Many modern organisms compartmentalize their respiration in dedicated cellular organoids, mitochondria. Their beginnings are beyond the scope of this article; let’s just stress that each mitochondrion is akin to a chemical reaction vessel where oxygen molecules are converted to water through several intermediate compounds. These intermediates are called reactive oxygen species because their reactivity is much higher than that of molecular oxygen and water. Such molecules can easily combine with DNA and oxidize it; the genetic material gets literally “rusty”. Proteins, lipids and other cellular components are also prone to oxidation, but they can be easily degraded and re-synthesized, while unique information-bearing molecules have to be repaired as soon as possible.
Scientists know several dozen oxidative DNA lesions, not all of them equally harmful. Some are encountered more frequently, others are less common; some cause mutations, others are innocuous... We’re about to meet perhaps the most malevolent one.
The wrong sort of letters
In the early 1980s, the Japanese government established a large grant program to investigate health effects of different ways to cook food. The Japanese, who traditionally eat many products uncooked (recall their famous sushi!), might be just suspicious about anything grilled or baked, or it could be that they are used to caring about their health. Regardless of the underlying reasons, this program allowed two chemists, Susumu Nishimura and Hiroshi Kasai, to do an experiment that at first looked as purely scientific curiosity. As the Russian writer Yuri Tomin put it, “What will happen if you take a brass kettle, pour in a glass of yoghurt there, add half a glass of kerosene, drop three ounces of ice cream and an old alarm clock, squeeze half a lemon, put a dead housefly, mix everything thoroughly, cover it with yesterday’s newspaper and zap it with X-rays?”
The Japanese chemists took a concentrated glucose solution, autoclaved it at 120 ° C, and got brown syrup, to which they then added one of DNA nucleobases, guanine. After many hours of heating this mixture, they separated it applying standard analytical techniques and looked for the products of guanine conversion. Glucose caramelizes when heated, as known to anyone who fancies homemade caramel. This complex reaction (actually, a bunch of concurrent reactions of eight different types!) produces hundreds of intermediates, some very reactive and capable of damaging DNA nucleobases. In their syrup, Kasai and Nishimura found two guanine derivatives, one of which was destined to infamy.
DNA molecules easily combine with reactive oxygen species.The genetic material literally rusts, like an iron bar left outdoors
Enter 8-oxoguanine. It is different from guanine only by two atoms, which do not even contribute to complementary bonding in normal DNA. However, the change in the configuration of atoms allows oxidized guanine to form a pair with A in addition to the normal pair with C. This causes the DNA polymerase to make errors when copying the strand containing 8-oxoguanine, inserting A opposite it. As a result, during the next round of copying, this A will, naturally, direct the incorporation of T at the place where G used to be.
This mutation, the replacement of G with T, belongs to a class of transversion mutations, which usually appear in DNA in the way just described. In some human genes, which trigger the development of cancer when mutated, transversions occur very frequently, and the oxidation of guanine in DNA is now considered an important carcinogenic mechanism. Mutations causing porphyria or hemophilia also often fall into this class.
At first, however, the report of the Japanese scientists did not arouse much interest: Who cares about the processes in the exotic model conditions of boiling syrup? Things changed a couple of years later, when in the USA Miral Dizdaroglu and Robert Floyd developed robust methods to detect vanishingly small quantities of 8-oxoguanine in DNA. Following that there began an incessant shower of papers addressing the chemistry and biology of this oxidized nucleobase.
It turned out that 8-oxoguanine forms when DNA is exposed to many environmental factors: ionizing radiation, sunlight, many chemical oxidants, tobacco smoke, motor engine exhaust, asbestos, etc. Not even a lead chamber with filtered air will provide a shelter: mitochondria in the cells of our own body spew out reactive oxygen species in abundance. Ideally, they all should be reduced to water, but no system in the physical world works ideally, and some of these very reactive molecules leak from mitochondria into the cell nucleus where they can damage DNA. What’s more, mitochondria are not the only source of dangerous oxidants in the organism. The human immune system, for example, employs special cells, called macrophages, which generate reactive oxygen species to kill pathogenic microorganisms.
So, even in the absence of environmental DNA-damaging factors, the DNA of each cell contains several thousand 8-oxoguanine nucleobases. Things get worse when the environment weighs in. An estimate made in the DNA of fish inhabiting the Seattle harbor, heavily polluted with industrial waste, revealed a whooping several-thousand-fold increase in the content of damaged nucleobases over the background. Nowadays, 8-oxoguanine is believed to be the most abundant of oxidative DNA lesions and is often used as a biomarker of oxidative stress because it is relatively easy to quantify.
Trackwalkers
It was clear from the beginning that such an important lesion should be repaired. The goal was to find the “trackwalker” enzyme that searches for 8-oxoguanine in DNA and removes it. Such proteins, called DNA glycosylases, had been known by that time for over a decade, but none of them could repair 8-oxoguanine.
In 1991, the laboratory of Arthur Grollman in the USA succeeded in the purification of the enzyme that could recognize 8-oxoguanine and excise it from DNA. This enzyme, purified from Escherichia coli bacterium, the workhorse of modern biology, was soon thereafter shown to be identical to another recently discovered protein, formamidopyrimidine-DNA glycosylase (Fpg for short), which excises yet another form of oxidized guanine.
Since then this enzyme has been actively studied in many laboratories over the world. In 2002, scientists from the Novosibirsk Institute of Bioorganic Chemistry (now the Institute of Chemical Biology and Fundamental Medicine, ICBFM), together with the Grollman group and their collaborators from the Hebrew University of Jerusalem, determined the three-dimensional structure of Fpg. To solve this task, which was labor-intensive and required exceptional technical skills, was necessary for understanding the enzyme’s mechanism, a prerequisite for any practical use.
In several laboratories abroad Fpg is currently tested as a reagent to protect healthy cells during cancer chemo- and radiotherapy. The Novosibirsk group looks into the role this enzyme plays in the life cycle of the bacterium that causes tuberculosis. Inhibition of some other glycosylases decreases the ability of the TB agent to resist human immune response, so in principle this phenomenon may pave the way to new drugs against this grave disease.
Even under normal conditions, the DNA of any single cell contains several thousand 8-oxoguanine molecules, making 8-oxoguanine one of the most abundant types of damage of genetic informationLet’s go back to the events inside a cell. The Fpg enzyme is highly specific: it can remove 8-oxoguanine that is paired with the “letter” C in DNA, but cannot do it when the lesion sits opposite A. This is quite logical since normal guanine is always complementary to C. If 8-oxoguanine is located opposite A, this can only mean that the DNA polymerase has already made a mistake when copying the damaged DNA strand; therefore, the removal of 8-oxoguanine from this position will immediately cause a mutation.
How do cells get around this trap? It turns out that in this case another DNA glycosylase comes into action. Called MutY, it recognizes the pair of 8-oxoguanine with A and, paradoxically, excises not the wrong but the normal “letter”, the adenine. Now the DNA polymerase can insert either C or A opposite 8-oxoguanine. If C is inserted, the problem is reduced to that already solved, and the repair is continued by Fpg protein. Even if the polymerase errs again and inserts A, there’s nothing to fear, as the MutY repair cycle can be repeated many times until C is finally inserted, and Fpg is engaged.
Fpg and MutY have a third “colleague”, an enzyme called MutT, which removes 8-oxoguanine from the pool of monomeric deoxynucleotide building blocks and prevents DNA polymerase from using the wrong sort of “letters” while making a new strand. It is of note that the names MutY and MutT are derived from mutation: the frequency of mutations is much higher in bacteria with the genes coding for these proteins “turned off”.
This three-tier system of 8-oxoguanine repair is so convenient that it is conserved in almost all live organisms, including us humans. Evidently, the enzymes can be structurally different from the bacterial ones and have different names, but the idea remains the same: remove 8-oxoguanine when paired with cytosine, excise adenine when paired with 8-oxoguanine.
In addition, there are enzymes that do not work on 8-oxoguanine but cleanse DNA of other oxidized bases. In E. coli, there are Nth and Nei proteins; in humans, NTH and several NEIL proteins. Similarly to Fpg, these components of the DNA repair system get a lot of attention from the scientists worldwide. Fortunately, Russia does not lag behind in this field, owing, to a large extent, to the efforts of scientists from Novosibirsk. A large number of studies are done at the ICBFM Laboratory of Repair Enzymes: a paper describing novel, previously unknown functions of the aforementioned NEIL proteins has just appeared; structures of several DNA glycosylases are in the pipeline; the researchers are actively collaborating with other laboratories at the ICBFM to study the dynamics of the enzyme structure and the role of DNA repair in the human body... Speakers from Novosibirsk are always welcome at all international meetings of scientists working in the field of DNA repair.
Feeling good?
What happens if our protein trackwalkers work sloppily? Possible results, as we have already seen, are mutations and tumor initiation due to the increased levels of 8-oxoguanine.
Human diseases arising from defects in the DNA repair system are extremely devastating; fortunately, they occur rarely. The best-known example is xeroderma pigmentodum, in which the repair of UV-induced lesions is defunct. The patients, numbered several thousand in the whole world, must be kept away from sunlight; once exposed, their skin gets blistered, malignant skin tumors develop. Most of these patients die young. Xeroderma, however, is caused by a failure of enzymes other than DNA glycosylases.
Until recently, no diseases have been known to be caused by mutations in the genes coding for DNA glycosylases. Moreover, knocking out these genes in laboratory mice did not lead to any adverse symptoms either. The situation became different in 2002, when a group from the University of Wales had reported the increased risk of colorectal cancer development in humans whose MutY gene carries a mutation. Nearly at the same time several groups showed that defects in the gene for uracil-DNA glycosylase — an enzyme that repairs non-oxidative lesions in DNA — resulted in severe immunodeficiency. Finally, a number of works on human 8-oxoguanine-DNA glycosylase confirmed the idea that some variants of this enzyme increased cancer risk in smokers and in people breathing polluted air.
Laboratory mice with a certain defect of DNA repair suffer from metabolic syndrome, a combination of obesity, hypertension and diabetesIn the beginning of 2006, the researchers from the University of Oregon created another line of knockout mice, this time with the NEIL1 gene inactivated. Again, the mutants didn’t run a greater risk of cancer than wild-type animals. Surprisingly, the scientists found that these mice were sick after all: They suffered from the so-called metabolic syndrome, a scourge of the First World population manifested in obesity, high blood pressure and diabetes. It is not known at present how the lack of NEIL1 can produce a phenotype never seen before for the defects in other DNA repair genes. Could it function in a small subset of cells in the body, for example, in the brain regions that regulate feeding behavior?
This is the kind of problems with which the researchers from Novosibirsk are involved, in collaboration or sometimes in intense competition with their colleagues from Russia and abroad. The “rusty” DNA is still an enigma.
Photographs by the author. The illustrations of three-dimensional structures made using PyMOL (DeLano Scientific).