Ancient DNA: From a Neanderthal to Sausage
In 2012, experts of the journal Science distinguished the work of a young German scientist, Matthias Meyer, who improved the protocol for preparing fossil DNA for an assay
Quite recently, nobody has believed that we will get to know more about the extinct animals and plants than their fossils can tell us. Indeed, paleontology gives much information about the body structure and even life style of extinct beings, but molecular genetic methods for assessing evolutionary relations, biochemistry, and physiology in this case seemed inapplicable.
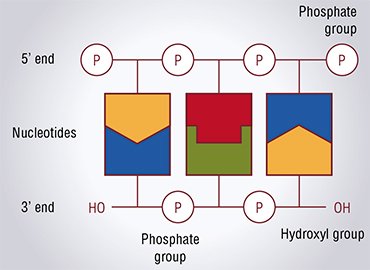 The analysis of ancient DNA, the field of biology that emerged literally right before our eyes during the last two decades, looks for many people committed to science as a miraculous fulfillment of a fairy dream. Although DNA is used in living nature as a major carrier of genetic information, its chemical stability is rather limited: DNA may be damaged by oxidation, hydrolysis, spontaneous loss of nucleotide bases, and so on. During the life of an organism, the specialized systems for DNA repair resist its damage; however, after death, DNA accumulates irreversible changes. In 1993, T. Lindal, a UK biochemist, published one of the most cited biological papers, where he demonstrated that DNA could not be preserved even under most favorable conditions (in the absence of oxygen and in permafrost) longer than one million years. If dead tissues are exposed to a high temperature or an aggressive chemical ambience, this period will be much shorter.
The analysis of ancient DNA, the field of biology that emerged literally right before our eyes during the last two decades, looks for many people committed to science as a miraculous fulfillment of a fairy dream. Although DNA is used in living nature as a major carrier of genetic information, its chemical stability is rather limited: DNA may be damaged by oxidation, hydrolysis, spontaneous loss of nucleotide bases, and so on. During the life of an organism, the specialized systems for DNA repair resist its damage; however, after death, DNA accumulates irreversible changes. In 1993, T. Lindal, a UK biochemist, published one of the most cited biological papers, where he demonstrated that DNA could not be preserved even under most favorable conditions (in the absence of oxygen and in permafrost) longer than one million years. If dead tissues are exposed to a high temperature or an aggressive chemical ambience, this period will be much shorter.
However, the technologies for assaying the vanishingly small DNA amounts based on PCR (polymerase chain reaction) and massively parallel sequencing have forged ahead too. These methods currently allow a complete determination of a DNA sequence from a single living cell, and specialists regard it trivial to obtain a sufficient amount of material from museum specimens or fossils whose age is not too ancient.
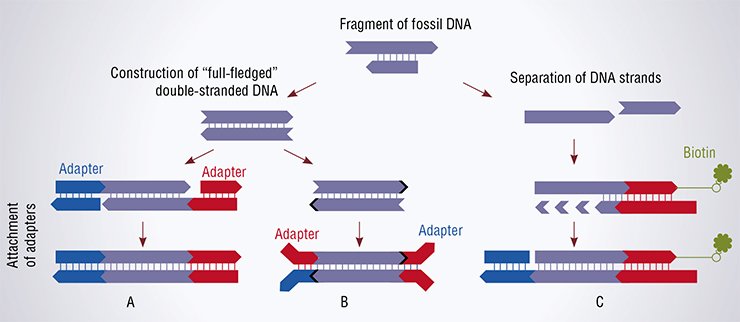
Correspondingly, both traditional methods start from treating a DNA wreck with the enzyme DNA polymerase, which, on the one hand, “reconstructs” the absent DNA strand formed of nucleotide bricks using overhanging 5’ ends as a template and, on the other hand, “eats away” the overhanging 3’ ends. The enzyme stops its work when all nucleotides are “paired” (according to the molecular biological slang, these ends are referred to as blunt ends).
Then the two traditional methods somewhat diverge yet retaining the main point—adapters (small synthetic double-stranded DNA with a known structure) are attached to a double-stranded ancient DNA fragment. Once all the manipulations have been completed, there is a double-stranded DNA molecule of an unknown sequence flanked by adapters; a set of such fragments is referred to as a DNA library, which is used for sequencing.
The main problem associated with using double-stranded DNA fragments for the construction of ancient DNA libraries is that the initial DNA is frequently rather significantly damaged. Thus, if one DNA strand carries several neighboring breaks in it, it just falls into pieces and is not included into the library. The DNA damages interfering with the work of DNA polymerase will lead to the same result. All this makes the sequencing accuracy for the ancient DNA rather low. When sequencing modern specimens, to exclude random errors, each position in the genome is independently read from different copies several tens of times (the so-called genome coverage). As for the published Denisovan genome, its genome coverage is only 1.9-fold and for the Neanderthal genome, only 1.3-fold, suggesting a high likelihood of error for any position of the sequence.
According to the technique elaborated by Meyer, the 5’-terminal phosphate group of the analyzed DNA is removed and then the DNA is heated to separate its strands. A single-stranded adapter, the molecule of biotin, is attached to the 3’ end of such single-stranded DNA. Biotin, better known as vitamin H or vitamin B7, serves as a kind of an anchor: it is able to stably bind to streptavidin, a bacterial protein, thereby making it possible to deposit such DNAs on the microbeads covered with this protein. Then the strand complementary to the adapter is added, and the enzyme DNA polymerase synthesizes the ancient DNA fragment using the available template. This gives a double-stranded fragment, which is supplemented with another pair of adapters.
The main advantage of this new technique is that each DNA fragment now has a twofold higher chance to get into the library. And taking into account that neither breaks nor other damages actually prevent adapters to bind with the analyzed DNA, representativeness of the library thus constructed increases severalfold rather than just doubles. The Denisovan human genome read with the help of Meyer’s method had a 30-fold coverage, exceeding that for the first contemporary human genome, sequenced only slightly over a decade ago
This is why scientists indulge us from time to time with news about deciphering the Neanderthal genome (which makes it possible to determine hair color, blood type, and even emotions) or about the possibility of restoring some irretrievably extinct species, such as the mammoth, dodo, or marsupial wolf. To read a genome has become a matter of technique; its synthesis will be feasible in the nearest future - it is just the issue of time and cost. The only problem to be solved will be to introduce this genome into the cell and to raise the embryo.
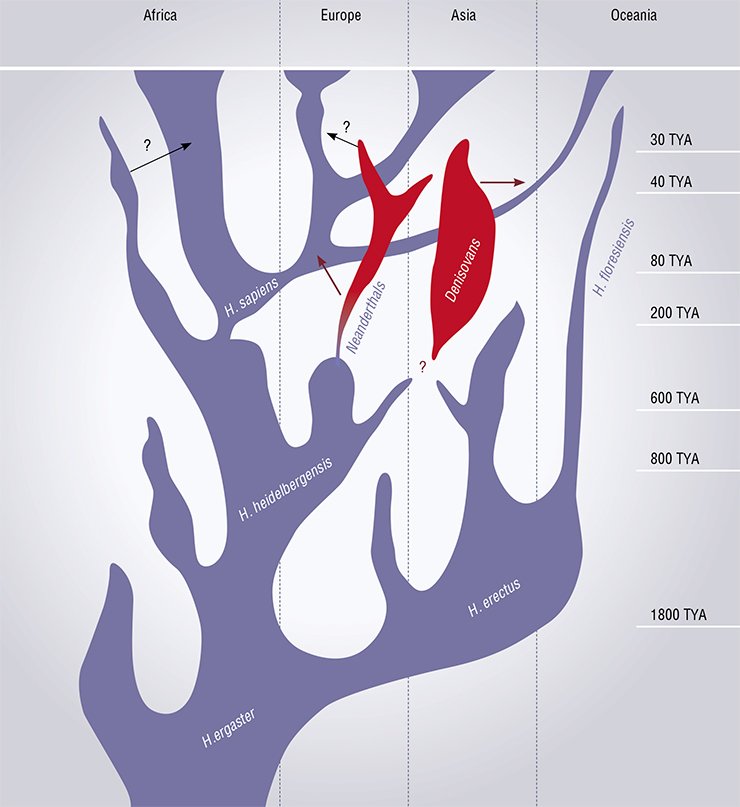
Not surprisingly, Science included the work by M. Meyer into the Breakthrough of the Year list; at the first glance, this work is far from revealing any basic problems. However, the experts of Science believe that the method by Meyer considerably advances the decoding of ancient DNA; moreover, the first application of this new technique is a comprehensive sequencing of the Denisovan human genome, a mysterious evolutionary branch of our ancestors discovered in Altai by the researchers of the Novosibirsk Institute of Archaeology and Ethnography, Siberian Branch, Russian Academy of Sciences.
Russian researchers do not stand aside from the current trends. In particular, specialists from different research institutions of the Siberian Branch of the Russian Academy of Sciences are involved in studying the Denisovans and animal fossils from the same Denisova Cave, or the experts in DNA repair at the Institute of Chemical Biology and Fundamental Medicine (Novosibirsk) try to adapt the corresponding enzymes for correcting the damages in ancient DNA before its sequencing.
By the way, similar problems are also encountered in applied fields, such as medicine (how to study the genome of a malignant tumor long ago excised from a patient and preserved in formalin), forensics (how to analyze the DNA of a piece of burnt bone), and even food industry (how to determine the raw stuff used to produce a sausage after all stages of deep processing).
Is it not a fairy dream to create the technology that would allow us both to determine the eye color of a Neanderthal individual and to catch a criminal?
The Editorial Board is grateful to M.V. Shun’kov, Doctor of History (Institute of Archaeology and Ethnography, Siberian Branch, Russian Academy of Sciences), for his assistance in preparing this paper